Family: Fimoviridae
Michele Digiaro, Toufic Elbeaino, Kenji Kubota, Francisco M. Ochoa-Corona and Susanne von Bargen
The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Michele Digiaro (digiaro@iamb.it)
Edited by: F. Murilo Zerbini, Sead Sabanadzovic and Luisa Rubino
Posted: August 2018, updated December 2023
PDF: ICTV_Fimoviridae.pdf
Summary
Members of the family Fimoviridae are plant viruses with a multipartite negative-sense single-stranded RNA genome (-ssRNA) composed of four to ten segments enveloped within spherical virions. They are distantly related to other ssRNA viruses (Elbeaino et al., 2018) (Table 1. Fimoviridae).
Table 1. Fimoviridae. Characteristics of members of the family Fimoviridae.
| Characteristic | Description |
Example
| fig mosaic virus (RNA1: AM941711; RNA2: FM864225; RNA3: FM991954; RNA4: FM992851; RNA5: HE803826; RNA6: HE803827) species Emaravirus fici |
| Virion | Approximately spherical and enveloped with a diameter of 80–150 nm |
| Genome | Four to ten segments of negative-sense ssRNA (12.3–18.5 kb in total) |
| Replication | Probably cytoplasmic |
| Translation | From capped mRNAs (produced by “cap snatching” from host mRNAs), which are complementary to the vRNAs |
| Host range | Mainly dicotyledonous species (camellia, Cercis, fig, grapevine, kiwi, lilac, oak, pear, pigeonpea, pistachio, raspberry, rose, rowan, etc.) and a few monocotyledonous species (wheat, maize, ti) |
| Transmission | Eriophyid mites. Some species are mechanically transmissible onto herbaceous indicators |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, subphylum Polyploviricotina, class Bunyaviricetes, order Elliovirales; one genus including 33 species |
Virion
Morphology
Virions are approximately spherical and enveloped with a diameter of 80–150 nm (Figure 1.Fimoviridae).
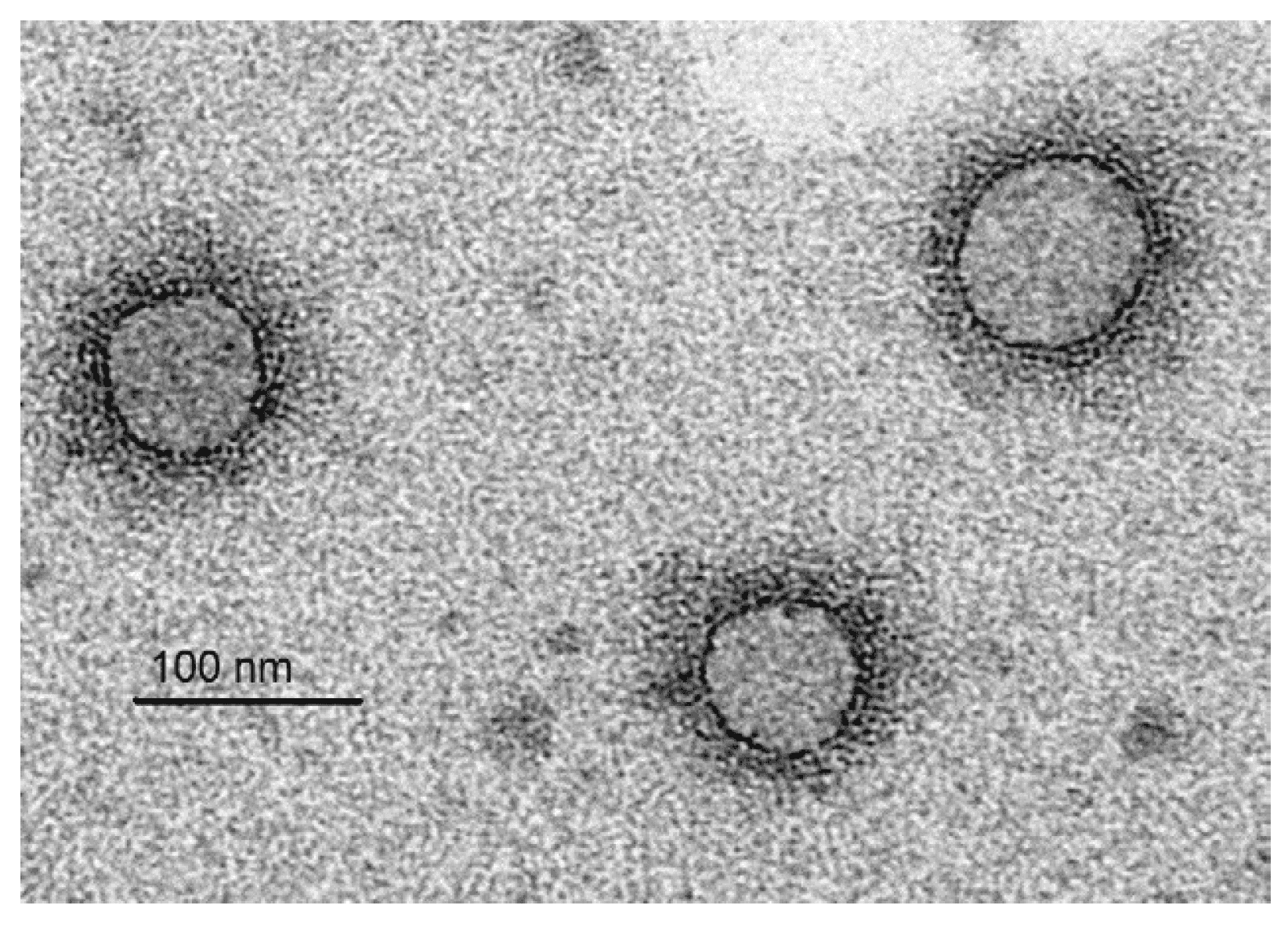 |
| Figure 1. Fimoviridae. Immunosorbent electron micrograph of virions of European mountain ash ringspot-associated virus. The bar represents 100 nm (Courtesy Inga Ludenberg, University of Hamburg, Germany). |
Physicochemical and physical properties
None reported.
Nucleic acid
The viral genome comprises from four segments of negative-sense ssRNA [palo verde broom virus (PVBV), Camellia japonica-associated virus 2 (CjaV2) and Ailanthus crinkle leaf-associated virus (ACrLaV)] to eight segments [raspberry leaf blotch virus (RLBV), and High Plains wheat mosaic virus (HPWMoV)], comprising 12.3 to 18.5 kb in total. A ninth additional RNA segment has been found in Camellia japonica-associated virus 1 (CjaV1), with RNA7 and RNA8 showing 53% amino acid identity, and in HPWMoV, which contains two RNA3 consensus sequences encoding the nucleocapsid protein (NP, 87.5% amino acid identity). Ten RNA segments have been observed in perilla mosaic virus (PerMV), which has two RNA3 segments (encoding the NP, 83% identity) and three RNA6 segments (29% to 63% amino acid identity) among them. Genomic RNAs are not capped or polyadenylated, and all contain complementary sequences (18–20 nt, depending on the RNA segment) at their 5′- and 3′-termini. The RNA-directed RNA polymerase (RdRP, RNA1) of fimovirids and its homolog (segment L, RNA1) in bunyaviral genomes share seven motifs (Pre-A, A-F) with conserved stretches of amino acid sequences (Elbeaino et al., 2009, Elbeaino et al., 2015, Kubota et al., 2021a).
Proteins
Each RNA segment of the genome encodes a single protein translated from the complementary strand as follows: RNA1 – RNA-directed RNA polymerase (RdRP, P1, 265–275.2 kDa); RNA2 – a glycoprotein precursor (GP, P2, 68.2–82.9 kDa) that is predicted to be cleaved into two products of 19.8–25.7 (Gn) and 50.9–53.1 kDa (Gc). A third smaller product of 2.6 kDa has been predicted in the case of AcCRaV and PerMV; RNA3 – the nucleocapsid protein (NP, P3, 29.5–36.0 kDa); RNA4 – a putative movement protein (MP, P4, ca. 35.4–43.6 kDa). The remaining RNAs (RNA5 to RNA9) encode proteins with unknown functions. The proteins encoded by non-core genome segments can be clustered in at least three main groups indicated as P55, ABC, and Glu2-Pro (Rehanek et al., 2022). A possible role of proteins encoded by RNA7 and RNA8 as suppressors of RNA silencing is hypothesized for HPWMoV (Gupta et al., 2019). On the other hand, ABC proteins are likely involved in pathogenicity, while a catalytic activity is suspected for Glu2-Pro (Rehanek et al., 2022).
Genome organization and replication
Fimoviruses differ from other related viruses (orthotospoviruses and orthobunyaviruses) by having a more significant number of genomic RNA segments. RNA1 to RNA4 encode each a single protein (Figure 2. Fimoviridae). The function of proteins encoded by the other RNAs (RNA5 to RNA9) remains unknown. In some fimoviruses two RNA3 consensus sequences encoding the NP have been found, as in the case of HPWMoV (Tatineni et al., 2014), chrysanthemum mosaic-associated virus (ChMaV) (Kubota et al., 2021b), PerMV (Kubota et al., 2020), and Pistacia virus B (PiVB) (Buzkan et al., 2019), with amino acid sequence divergence ranging from 12.5% to 52%. Also, non-structural proteins encoded by additional RNAs of some fimoviruses seem to be present as multiple copies within the genome. This fact has been shown, for instance, for the P5 and P6 of European mountain ash ringspot-associated virus (EMARaV) (von Bargen et al., 2019), the P5 of PiVB (Buzkan et al., 2019) and the P6 of PerMV (Kubota et al., 2020), while RNA4 and RNA5 of jujube yellow mottle-associated virus (JYMaV) encode two MPs (Yang et al., 2019). The significance of these protein duplicates and their function remains to be clarified. Like other multipartite -ssRNA viruses, some fimoviruses use the “cap snatching” to initiate transcription and facilitate translation of their mRNAs, as reported for rose rosette virus (RRV) and fig mosaic virus (FMV) (Laney et al., 2011, Walia and Falk 2012). Cap-snatching involves binding of host-capped mRNAs to the ribonucleoproteins (RNPs), cleavage of these RNAs close to the 5′ cap by a viral endonuclease activity, and use of the short-capped fragments as primers for viral mRNA transcription. As for other bunyaviruses, the replication of fimoviruses probably occurs in the cytoplasm.
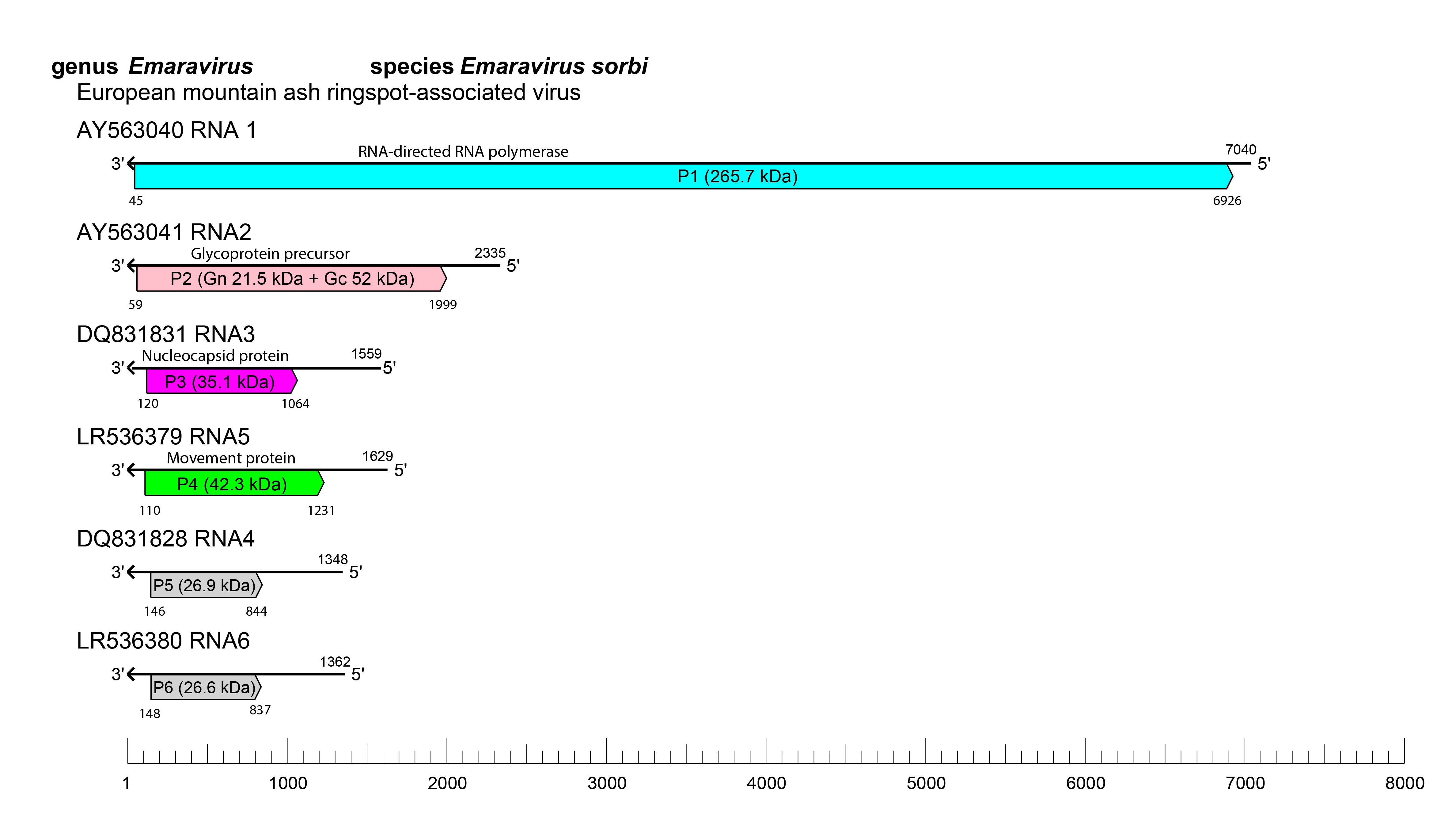 |
| Figure 2. Fimoviridae. Schematic representation of the organization of the six RNA segments constituting the genome of EMARaV. ORFs are represented as coloured boxes with annotation indicating each protein's function and estimated molecular mass. Grey color indicates ORFs encoding proteins of unknown function. |
Biology
Except for HPWMoV, which infects wheat and maize, and ti ringspot-associated virus (TiRSaV), which infects ti plant, the other fimoviruses almost exclusively infect dicotyledons, mainly forest (ash, aspen, karaka, maple, mountain ash, oak, palo verde, redbud) and cultivated woody species (fig, grapevine, Japanese star anise, jujube, pear, kiwi, pistachio), but also numerous shrub species (blackberry, pigeonpea, Rubus spp.), herbaceous species (alfalfa, pea, perilla) and ornamentals (camellia, chrysanthemum, cyclamen, lilac, rose).
Depending on the host plant, symptoms of fimovirus infection vary from leaf mottling, blotching, vein clearing, chlorotic ringspots, yellowing, and sterility of flowers to a general mosaic. Fimoviruses are transmitted by grafting, and many have been proven to be vectored by eriophyid mites (Epstein and Hill 1995, Kumar et al., 2003, Elbeaino and Digiaro 2021). The mechanical transmission of fimoviruses onto herbaceous hosts is rare; it has been shown only for a few species, i.e., AcCRaV, pigeonpea sterility mosaic virus 1 (PPSMV1), pigeonpea sterility mosaic virus 2 (PPSMV2), LiCRaV, PerMV, Pueraria lobata-associated emaravirus (PloaEV), RLBV, RRV and TiRSaV.
Derivation of names
Fimoviridae: from fig mosaic virus
Emaravirus: from European mountain ash ringspot-associated virus.
Genus demarcation criteria
The family includes only a single genus. Based on aspects that consider phylogeny, encoded proteins, and genomic terminations, the possibility of dividing the genus Emaravirus into at least two distinct genera should be considered in a future taxonomic proposal (Rehanek et al., 2022).
Relationships within the family
Phylogenetic trees constructed with amino acid sequences of RdRP (Figure 3. Fimoviridae), GP, NP, or MP of the fimoviruses show similar clustering with four main groups. The first group (I) includes Actinidia virus 2 (AcV2), ash shoestring-associated virus (ASaV), fig mosaic virus (FMV), Pueraria lobata-associated emaravirus (PloaEV)*, pigeonpea sterility mosaic virus 2 (PPSMV2), aspen mosaic-associated virus (AsMaV), Pistacia virus B (PiVB), maple mottle-associated virus (MaMaV), rose rosette virus (RRV), blackberry leaf mottle-associated virus (BLMaV), pigeonpea sterility mosaic virus 1 (PPSMV1), Vitis emaravirus (VEV), European mountain ash ringspot-associated virus (EMARaV), Actinidia chlorotic ringspot-associated virus (AcCRaV), lilac chlorotic ringspot-associated virus (LiCRaV), and redbud yellow ringspot-associated virus (RYRSaV). The second group (II) includes palo verde broom virus (PVBV), Callicarpa mosaic-associated virus 1 (CMaV1)*, High Plains wheat mosaic virus (HPWMoV), Ailanthus crinkle leaf-associated virus (ACrLaV)*, common oak ringspot-associated virus (CORaV), ti ringspot-associated virus (TiRSaV), Arceuthobium sichuanense-associated virus 1 (ArSaV1), raspberry leaf blotch virus (RLBV), and jujube yellow mottle-associated virus (JYMaV). The third group (III) includes pear chlorotic leaf spot-associated virus (PCLSaV), Callicarpa mosaic-associated virus 2 (CMaV2)*, chrysanthemum mosaic-associated virus (ChMaV), and karaka Õkahu purepure emaravirus (KOPV). The fourth group (IV) includes Camellia japonica-associated virus 1 (CjaV1), Camellia japonica-associated virus 2 (CjaV2), Japanese star anise ringspot-associated virus (JSARaV), Artemisia fimovirus 1 (ArtV1)*, and perilla mosaic virus (PerMV). Viruses with asterisks (*) are putative fimoviruses that have yet to be assigned to a species in the family.
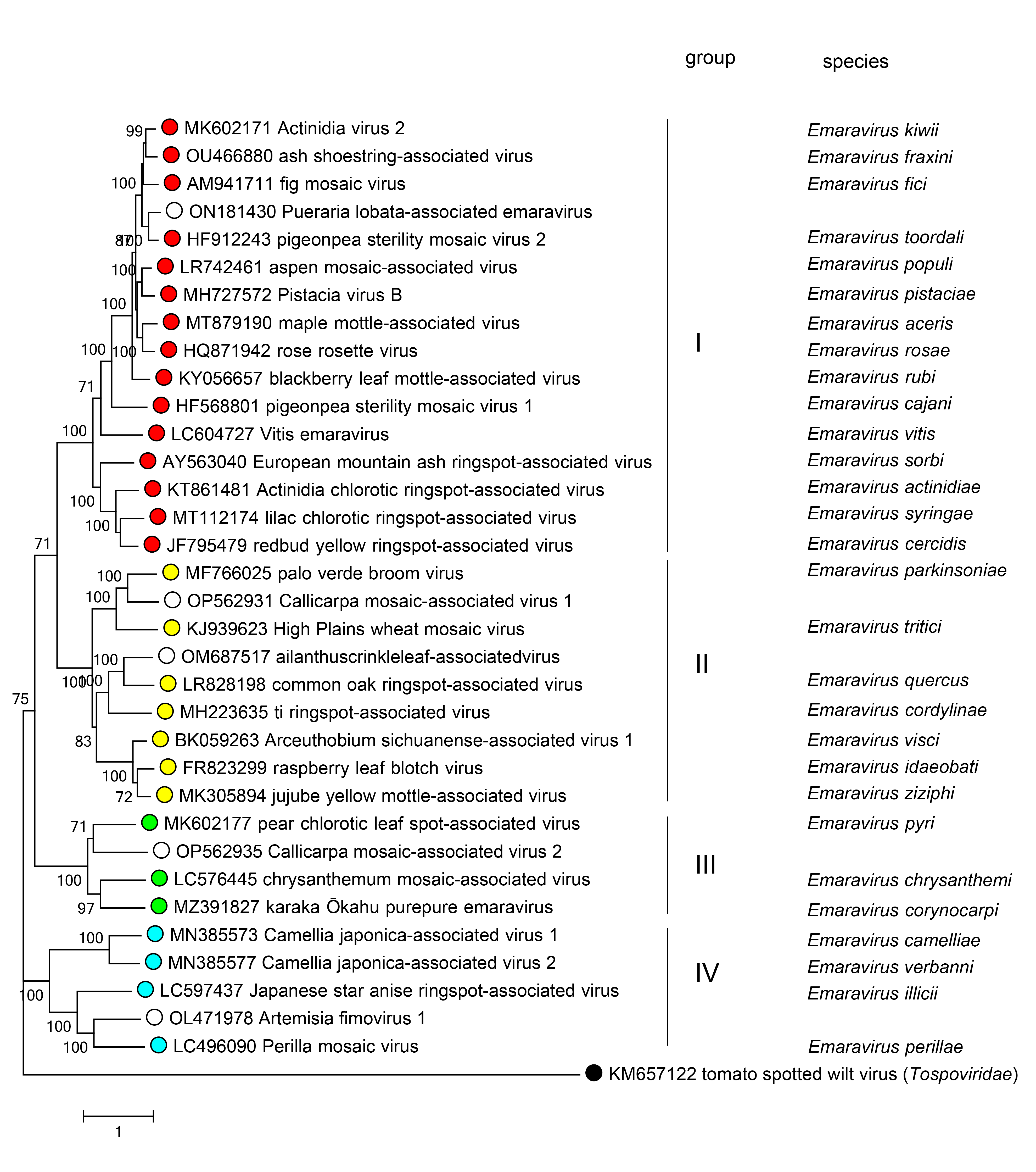 |
| Figure 3. Fimoviridae. Phylogenetic tree constructed with amino acid sequences encoded by RNA1 (RdRP) of members of the family Fimoviridae, using MUSCLE alignment and Maximum Likelihood method implemented in MEGA11 (Tamura et al., 2013). The GenBank accession numbers of the sequences used are reported next to the virus names. Bootstrap values above 50% are shown at branch points, with 1000 replicates. Viruses with white circles are putative fimoviruses of the family Fimoviridae that still need to be officially recognized. Tomato spotted wilt virus was used as an outgroup species. |
Relationships with other taxa
Viruses of the family Fimoviridae (order Elliovirales) are related to those of the families Tospoviridae and Peribunyaviridae in that they share (i) a multipartite -ssRNA genome of four to ten segments; (ii) high sequence identity with orthologous proteins of members of the order Elliovirales at equivalent genome positions in the first three RNAs (corresponding to L, M, and S RNA segments), i.e. RNA-directed RNA polymerase (RdRP, RNA1), putative glycoprotein precursor (GP, RNA2) and putative nucleocapsid protein (NP, RNA3); (iii) seven conserved motifs (preA-A-F) in the amino acid sequence of their RdRP; (iv) enveloped virions; (v) stretches of nucleotides at both 5′- and 3′-termini of all RNA segments that are almost perfectly complementary to each other. These terminal sequences have several nucleotides conserved in all genomic RNAs of fimoviruses, and are similar, but not identical, to those of other members of the order Elliovirales.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference |
| alfalfa ringspot-associated virus | RNA1: MK648429*; RNA3: MK648430* | AlRSaV | (Samarfard et al., 2020) |
| Ailanthus crinkle leaf-associated virus | RNA1: OM687517; RNA2: OM687518; RNA3: OM687519; RNA4: OM687520 | ACrLaV | (An et al., 2022) |
| Artemisia fimovirus 1 | RNA1: OL471978; RNA2: OL471979; RNA3: OL471980; RNA4: OL471981; RNA5: OL471982; RNA7: OP441764 | ArtV1 | (Rivarez et al., 2023) |
| Callicarpa mosaic-associated virus 1 | RNA1: OP562931; RNA2: OP562932; RNA3: OP562933; RNA4: OP562934 | CMaV1 | |
| Callicarpa mosaic-associated virus 2 | RNA1: OP562935; RNA2: OP562936; RNA3: OP562937; RNA4: OP562938 | CMaV2 | |
| Pueraria lobata-associated emaravirus | RNA1: ON181430; RNA2: ON181431; RNA3: ON181432; RNA4: ON181433; RNA5: ON181434 | PloaEV |
|
| woolly burdock yellow vein virus | RNA3: JQ354894* | WBYVV
| (Bi et al., 2012) |
| Yunnan emara-like virus | RNA1: MW896848*; RNA2: MW896849*; RNA3: MW896850*; RNA4: MW896851*; RNA5: MW896852; RNA6: MW896853; RNA8: MW896854 | YEV | (Chen et al., 2022) |
* Partial sequences
Virus names and virus abbreviations are not official ICTV designations.

