Family: Endornaviridae
Rodrigo A. Valverde, Mahmoud E. Khalifa, Ryo Okada, Toshiyuki Fukuhara and Sead Sabanadzovic
The citation for this ICTV Report chapter is the summary published as Valverde et al., (2019):
ICTV Virus Taxonomy Profile: Endornaviridae, Journal of General Virology, 100, 1204–1205.
Corresponding author: Rodrigo A. Valverde (rvalverde@agcenter.lsu.edu)
Edited by: F. Murilo Zerbini and Sead Sabanadzovic
Posted: April 2019, updated June 2020
PDF: ICTV_Endornaviridae.pdf
Summary
The family Endornaviridae includes viruses with linear, single-stranded, positive-sense RNA genomes that range from 9.7 to 17.6 kb and have been reported infecting plants, fungi, and oomycetes (Table 1. Endornaviridae). The family consists of two genera, Alphaendornavirus and Betaendornavirus, into which viruses are classified based on their genome size, host, and presence of unique domains. Alphaendornavirus includes 24 species whose members infect plants, fungi and oomycetes, while the genus Betaendornavirus includes 7 species whose members infect ascomycete fungi.
Table 1. Endornaviridae. Characteristics of members of the family Endornaviridae.
| Characteristic | Description |
| Typical member | Oryza sativa endornavirus Nipponbare (D32136), species Alphaendornavirus oryzae |
| Virion | No true virions are associated with members of this family |
| Genome | Monocistronic single-stranded positive-sense RNA of 9.7–17.6 kb |
| Replication | Cytoplasmic |
| Translation | From monocistronic positive-sense RNA |
| Host range | Host-specific; plants, fungi and oomycetes |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Kitrinoviricota, class Alsuviricetes, order Martellivirales; two genera including 31 species |
Virion
Morphology
No true virions are associated with members of this family since their genomes lack a coat protein gene. However, their RNA genomes have been associated with pleomorphic cytoplasmic membrane vesicles. Well-defined cytoplasmic vesicles of ~70 nm diameter have been reported in the cytoplasm of the '447' male sterile strain of Vicia faba (broad bean) infected with Vicia faba endornavirus (VfEV) (Lefebvre et al., 1990, Dulieu et al., 1988).
Nucleic acid
Viruses in the family Endornaviridae consist of a single molecule of naked positive-sense RNA of 9.7 to 17.6 kb (Fukuhara and Gibbs 2012, Stielow et al., 2011). Most endornaviruses have been characterized using the viral replicative forms (dsRNAs), which are relatively stable, present in relatively high quantities in the host tissue, and easily isolated (Figure 1. Endornaviridae.). The terminal sequence of the 3′-end of most endornavirus RNAs consists of short stretch of repeated cytosine residues. The dsRNA of several plant endornaviruses contains a site-specific break (nick) in the coding strand at about 1.2–2.7 kb from the 5′-terminus (Fukuhara et al., 1995, Okada et al., 2011, Okada et al., 2013).
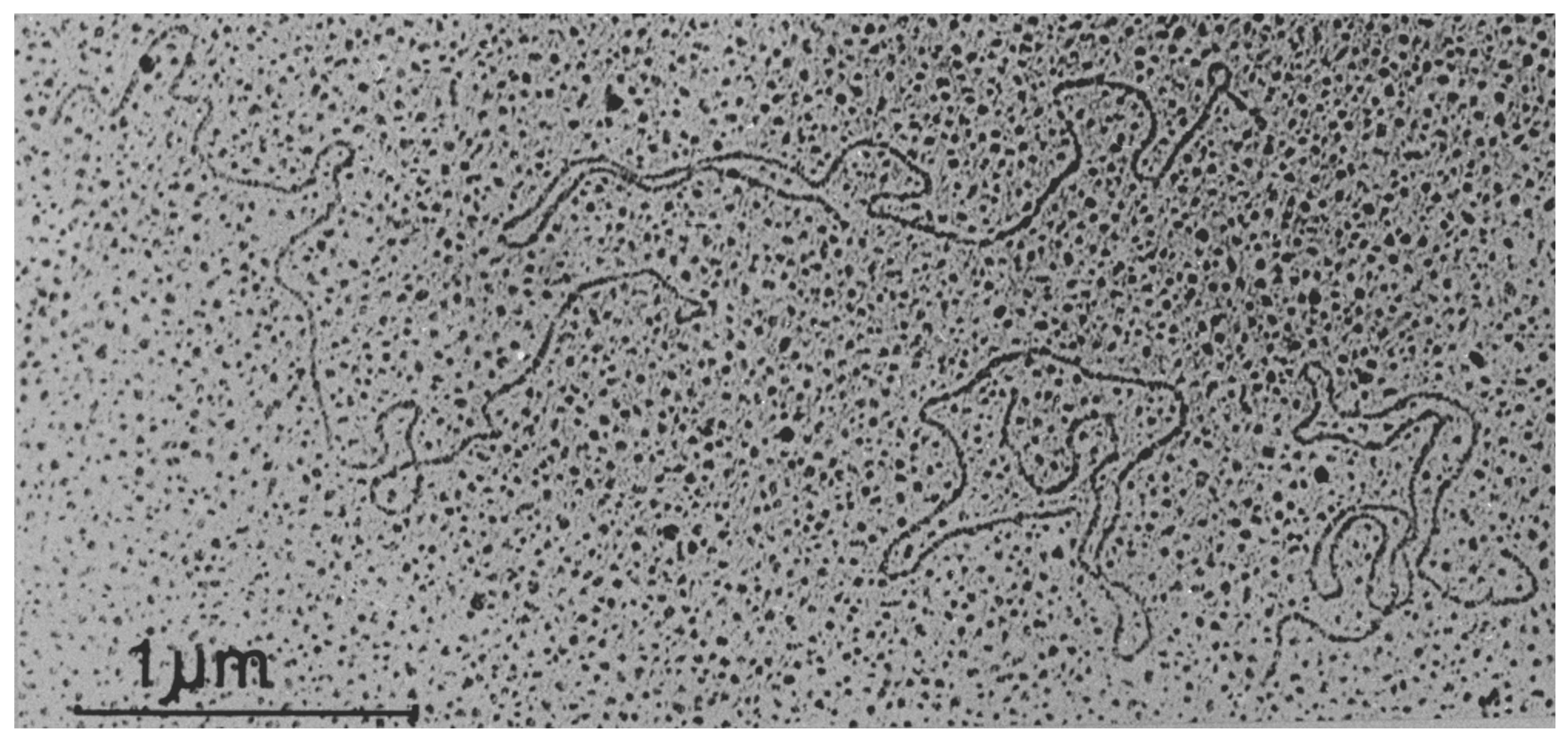 |
| Figure 1. Endornaviridae. Transmission electron micrograph of dsRNA molecules of Oryza sativa endornavirus isolate Nipponbare. Figure reproduced with permission (Fukuhara et al., 1993). Bar represents 1 µm. |
Proteins
The RNA of viruses belonging to the Endornaviridae is not associated with structural proteins.
Genome organization and replication
The genome of endornaviruses consists of a linear ssRNA containing a single ORF encoding a polyprotein that ranges from 3,217–5,825 aa (Fukuhara and Gibbs 2012, Stielow et al., 2011, Gibbs et al., 2000). A conserved RNA-directed RNA polymerase (RdRP) domain located in the C-terminal region of the polyprotein is a common feature among all endornaviruses. Other functional domains found in some, but not all, endornaviruses include viral helicase superfamily 1, methyl transferase, glycosyl transferase, cysteine-rich region, phytoreo_S7 domain, and capsular polysaccharide synthase (Okada et al., 2011, Okada et al., 2013, Hacker et al., 2005, Sabanadzovic et al., 2016). Several other, still uncharacterized, proteins are likely encoded by the large ORFs of endornaviruses (Fukuhara and Gibbs 2012), including one or more proteinases to cleave the encoded polyprotein precursor into several functional proteins.
RNA-directed RNA polymerase activity associated with Oryza sativa endornavirus (OsEV) has been detected in the crude microsomal fraction of rice cultured cells and with cytoplasmic vesicles containing VfEV RNA (Lefebvre et al., 1990, Horiuchi et al., 2001). There is evidence that endornaviruses may encode RNA silencing suppressors to counteract the host's antiviral silencing machinery (Fukuhara and Gibbs 2012, Sela et al., 2012). Endornavirus-derived small RNAs (siRNAs) have been detected in plants infected with OsEV (Urayama et al., 2010), bell pepper endornavirus (BPEV) (Sela et al., 2012), Phaseolus vulgaris endornavirus 1 (PvEV1) and Phaseolus vulgaris endornavirus 2 (PvEV2) (Nordenstedt et al., 2017) indicating that the hosts’ RNA silencing machinery recognizes the endornavirus RNA. The genomes of BPEV, PvEV1 and PvEV2 have been successfully assembled from siRNAs using high throughput sequencing. Knock-down of the dicer-like 2 gene in rice negatively affects maintenance of OsEV (Urayama et al., 2010).
Biology
Endornaviruses have been reported in several economically important crops and their wild relatives, in plant pathogenic fungi, and in species of the oomycete Phytophthora. Because of their persistent lifestyle and lack of effect on the host phenotype (with a couple of exceptions detailed elsewhere in this chapter), little is known about the biology of these viruses. However, endornaviruses may have significant effects on the evolution of acute viruses as well as their hosts (Roossinck 2010).
Several studies have shown that the dsRNA of plant endornaviruses is localized in the cytoplasm (about 100 copies per cell) (Moriyama et al., 1996). Plant endornaviruses are distributed throughout all tissues and are transmitted only through the gametes. Indirect evidence suggests that, in plants, endornaviruses lack cell-to-cell movement and this is the reason for the lack of transmissibility by conventional methods (Valverde and Gutierrez 2007). However, they are efficiently transmitted via seeds (Okada et al., 2011, Valverde and Gutierrez 2007, Horiuchi et al., 2003) and egg cell and pollen (Moriyama et al., 1996, Horiuchi et al., 2003, Moriyama et al., 1999), probably due to the 10-fold increase in their copy number in pollen grains. Fungal endornaviruses, like many mycoviruses, are efficiently transmitted vertically through spores (Park et al., 2006) and horizontally via hyphal anastomosis (Ikeda et al., 2003).
Two endornaviruses have been associated with phenotypic changes in their host, the plant endornavirus VfEV which is associated with cytoplasmic male sterility in the Vicia faba line '447' (Lefebvre et al., 1990) and the fungal endornavirus, Helicobasidium mompa endornavirus 1 (HmEV1), associated with the hypovirulence trait in strain V670 of the violet root rot fungus, Helicobasidium mompa (Osaki et al., 2006). Comparative studies aiming at the evaluation of the plant phenotype of endornavirus-infected and endornavirus-free lines, using common bean (Phaseolus vulgaris) plants as a study model, have not yielded detectable differences, suggesting that endornaviruses are not associated with visible pathogenic effects (Khankhum and Valverde 2018). However, variations in physiological traits have been reported in the same study. Indeed, the endornavirus-infected lines were significantly different than those free of the virus in having faster seed germination, longer radicles, lower chlorophyll content, higher carotene content, longer pods, and higher seed weight (Khankhum and Valverde 2018). Analysis of RNAseq data obtained from endornavirus-infected (PvEV1 and PvEV2) and endornavirus-free common bean plants identified 132 differentially expressed genes, most of which are associated with oxidation-reduction processes (Khankhum et al., 2016). In the case of fungal endornaviruses, in most cases the endornavirus has no obvious effect on either host phenotype or virulence (Khalifa and Pearson 2014).
Antigenicity
Serological studies of endornaviruses are limited to VfEV. Monoclonal antibodies against purified cytoplasmic vesicles from plants infected with VfEV have been generated and used to detect the virus. The antibodies recognized an epitope in the VfEV-associated vesicles that contains sugars, possibly a glycolipid (Dulieu et al., 1988).
Derivation of names
Endorna: from endo, Greek meaning "within", and RNA.
Most endornaviruses have been named after the common or botanical name of the host.
Genus demarcation criteria
The demarcation criteria for the two genera of the Endornaviridae are:
- Genome length (>11.9 kb for genus Alphaendornavirus and <10.7 kb for genus Betaendornavirus).
- Separate phylogenetic clustering of RdRP sequences from viruses in each genus.
- The presence/absence of a site-specific nick near the 5′-end of the coding strand (present in most members of Alphaendornavirus, but absent in members of Betaendornavirus).
Relationships within the family
Phylogenetic analysis of RdRP aa sequences from different endornaviruses reveals that they cluster into two well-separated clades corresponding to the two genera in the family (Figure 2. Endornaviridae).
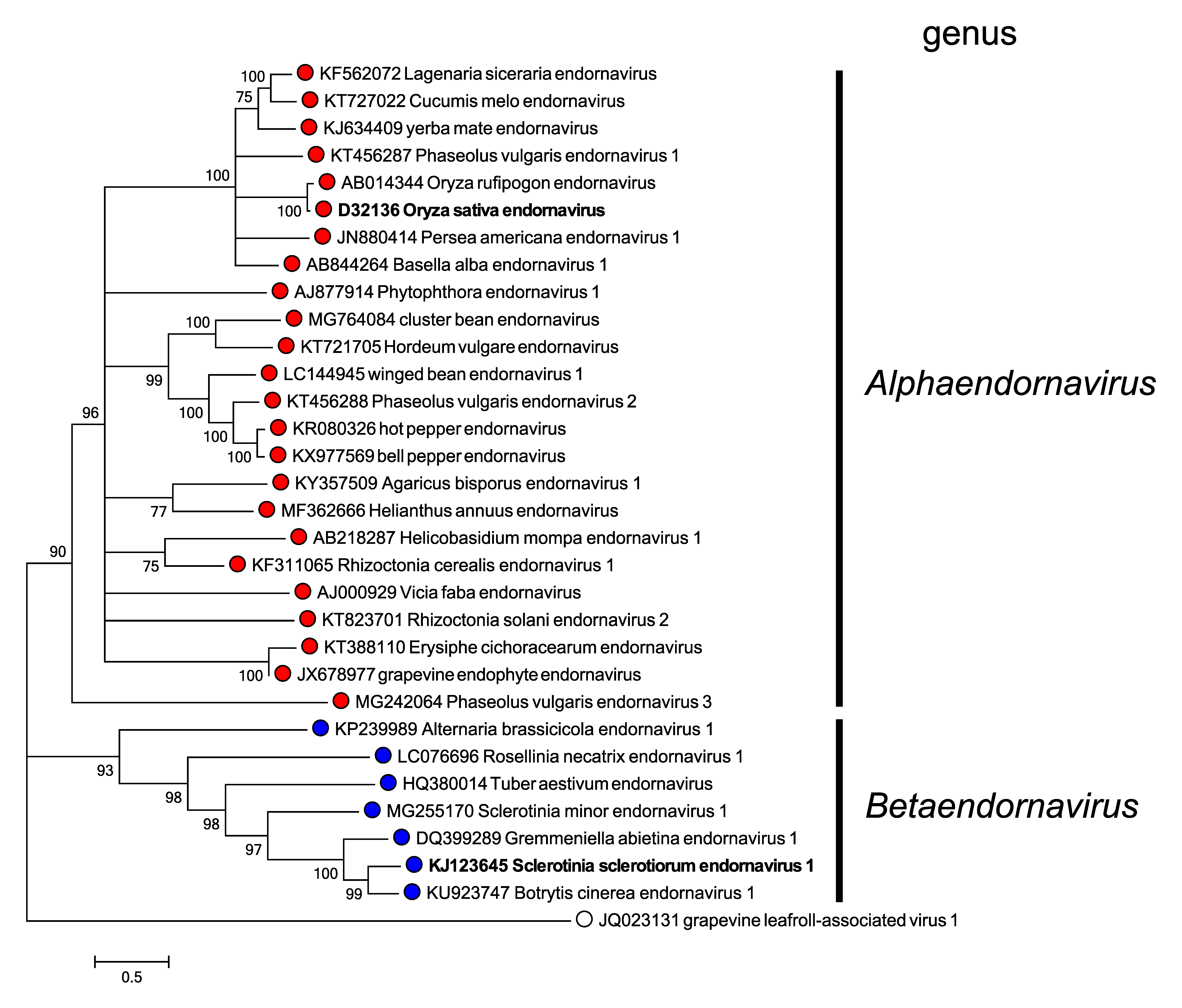 |
| Figure 2. Endornaviridae. Maximum likelihood phylogenetic tree constructed using the amino acid sequences of RNA-directed RNA polymerase domains of endornaviruses. Grapevine leafroll-associated virus 1 (genus Ampelovirus, family Closteroviridae) was used as an outgroup. For each genus, a virus belonging to the type species is indicated by bold text. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
The RdRP and helicase domains of endornaviruses are related to those of the "alpha-like" superfamily of positive-sense RNA viruses (Gibbs et al., 2000).
Evolutionary analyses of endornavirus genomes revealed that several of them contain regions that are homologous to plant, fungal and bacterial genes, suggesting their acquisition via lateral gene transfer (Sabanadzovic et al., 2016, Roossinck et al., 2011, Song et al., 2013). For instance, the glycosyltransferase 28 domain of BPEV, as well as the glycosyltransferase sugar-binding domain and a capsular polysaccharide synthesis protein of OsEV and CmEV, may have originated from bacteria. Moreover, it is hypothesized that lateral gene transfer has a role in the origin of the S7 domain present in several endornaviruses (Khalifa and Pearson 2014, Liu et al., 2012).
Related, unclassified viruses
Unclassified endorna-like viruses have been identified in different plant and fungal hosts. The molecular and biological properties of these viruses are similar to those of recognized members of the Endornaviridae.
| Virus name | Accession number | Virus abbreviation |
| balloon flower endornavirus | MN015676 | BfEV |
| Ceratobasidium endornavirus A | KX355142 | CbEVA |
| Ceratobasidium endornavirus B | KX355143 | CbEVB |
| Ceratobasidium endornavirus C | KX355164 | CbEVC |
| Ceratobasidium endornavirus D | KX355144 | CbEVD |
| Ceratobasidium endornavirus E | KX355145 | CbEVE |
| Ceratobasidium endornavirus F | KX355146 | CbEVF |
| Ceratobasidium endornavirus G | KX355147 | CbEVG |
| Ceratobasidium endornavirus H | KX355148 | CbEVH |
| Chalara elegans endornavirus 1 | GQ494150 | CeEV1 |
| Diplodia seriata endornavirus 1 | MK584822 | DsEV1 |
| Gyromitra esculenta endornavirus 1 | MK279476 | GeEV1 |
| Morchella importuna endornavirus 1 | MK279476 | MiEV1 |
| Morchella importuna endornavirus 2 | MK279478 | MiEV2 |
| Morchella importuna endornavirus 3 | MK279479 | MiEV3 |
| Rhizoctonia solani endornavirus | KC792590 | RsEV |
| Sclerotium rolfsii endornavirus 1 | MH766493 | SrEV1 |
Virus names and virus abbreviations are not official ICTV designations.

