Family: Druskaviridae (Interim Report)
This is a summary page created by the ICTV Report Editors using information from associated Taxonomic Proposals and the Master Species List.
Edited by: Mart Krupovic
Posted: June 2023, updated December 2024
Summary
The family Druskaviridae includes dsDNA viruses infecting hyperhalophilic archaea of Haloarcula sp. (Table 1 Druskaviridae). A characteristic feature of viruses in this family is that they encode the preQ0/G+ pathway responsible for the formation of 7-deazaguanine modification in the genomic DNA. The family Druskaviridae was established in 2022 (Master Species List 37).
Table 1 Druskaviridae. Characteristics of members of the family Druskaviridae.
| Characteristic | Description |
| Example | Haloarcula vallismortis tailed virus 1 (KC117377), species Tredecimvirus thailandense, genus Tredecimvirus |
| Virion | Icosahedral head of approximately 85 nm diameter and long noncontractile flexible tail (siphovirus-like morphology). |
| Genome | Linear, dsDNA genome of 102–104 kbp, direct terminal repeats (Figure 1 Druskaviridae) |
| Host range | Hyperhalophilic archaea of Haloarcula sp. |
| Realm Duplodnaviria, kingdom Heunggongvirae, phylum Uroviricota, class Caudoviricetes, order Thumleimavirales: 2 genera and 2 species (Figure 2 Druskaviridae) |
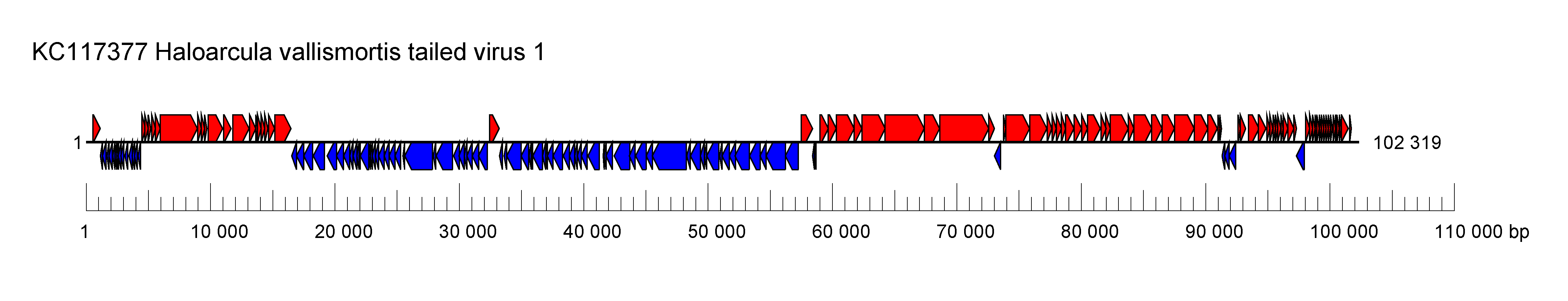 |
| Figure 1 Druskaviridae. Genome organisation of Haloarcula californiae tailed virus 1, a member of the family Druskaviridae. Boxes indicate open reading frames as annotated on GenBank accession KC117377. |
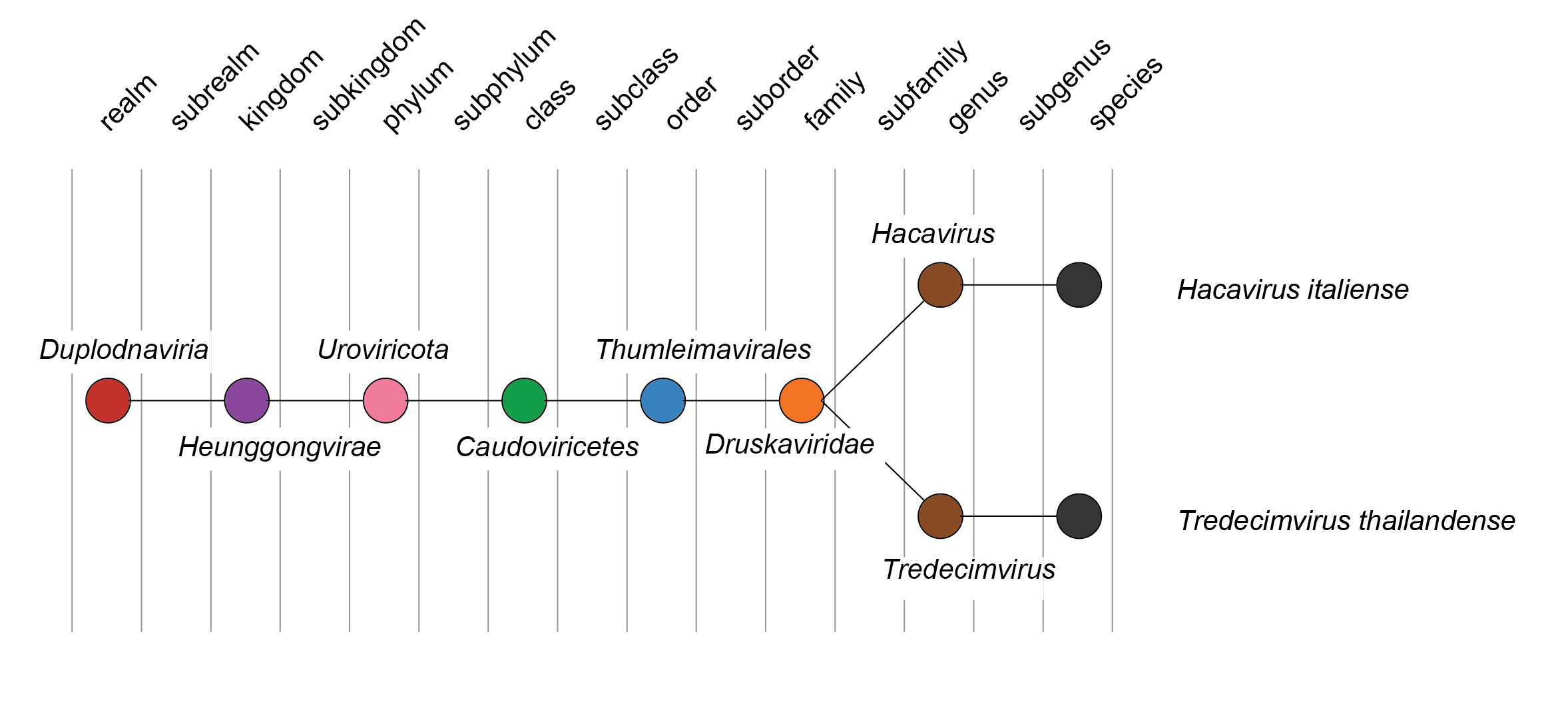 |
| Figure 2 Druskaviridae. Taxonomy of the family Druskaviridae. |
Derivation of name
Druskaviridae: From Lithuanian druska, for salt

