Family: Corticoviridae
Hanna M Oksanen
The citation for this ICTV Report chapter is the summary published as Oksanen et al., (2017):
ICTV Virus Taxonomy Profile: Corticoviridae, Journal of General Virology, 98, 888–889.
Corresponding author: Hanna M. Oksanen ([email protected])
Edited by: Andrew M. Kropinski and Andrew J. Davison
Posted: June 2017, updated June 2023
PDF: ICTV_Corticoviridae.pdf (2017 version)
Summary
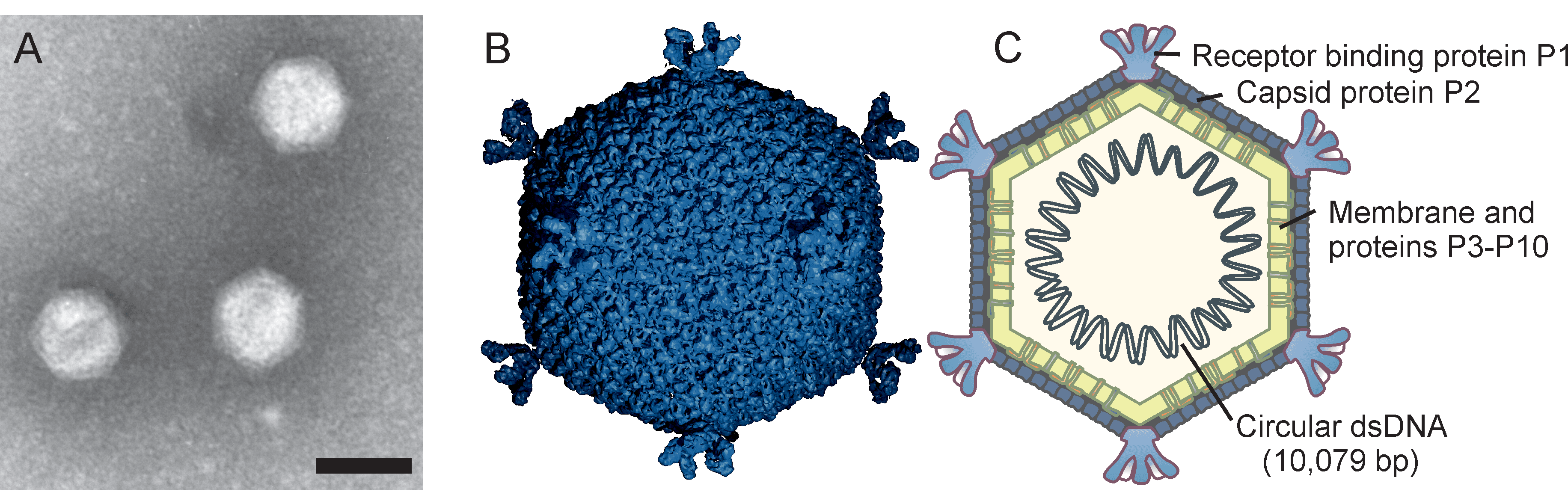
Corticoviridae is a family of icosahedral, internal membrane-containing viruses with double-stranded circular DNA genomes of approximately 10 kbp (Table 1 Corticoviridae). Two species, Merivirus PM2, and Merivirus Cr39582 are recognized. Pseudoalteromonas virus PM2 (PM2) infects gram-negative bacteria and was isolated from seawater in 1968 and was the first bacterial virus in which the presence of lipids in the virion was demonstrated. Viral lipids are acquired selectively during virion assembly from the host cytoplasmic membrane. The outer protein capsid is an icosahedron with a pseudo T=21 symmetry and an internal protein-rich membrane enclosing the genome.
Table 1.Corticoviridae. Characteristics of members of the family Corticoviridae.
| Characteristic | Description |
| Example | Pseudoalteromonas virus PM2 (AF155037), species Merivirus PM2 |
| Virion | Icosahedral, internal membrane-containing virions of about 57 nm with a single capsid protein P2, single spike protein P1 and 8 membrane-associated proteins P3–P10 |
| Genome | 10.1 kb of highly supercoiled, circular, double-stranded DNA |
| Replication | Rolling circle replication initiated by virus-encoded protein P12 |
| Translation | Prokaryotic translation using viral mRNA and host ribosomes |
| Host Range | Bacteria, gram-negative Pseudoalteromonas strains |
| Taxonomy | Realm Varidnaviria, kingdom Abadenavirae, phylum Produgelaviricota, class Belvinaviricetes, order Vinavirales; one genus including two species |
Virion
Morphology
Knowledge of virion morphology is based on studies of the PM2 phage. Icosahedral virions consist of an internal membrane and an outer protein capsid that has a diameter of 57 nm between facets (Figure 1 Corticoviridae) (Abrescia et al., 2008). The genome is enclosed by the membrane. The capsid consists of 200 major capsid protein P2 trimers that are organized on a pseudo T=21 lattice (Abrescia et al., 2008). Protein P2 is composed of two beta-barrels disposed normal to the capsid surface. The P2 trimers have pseudo-six-fold symmetry and the structure is stabilized by calcium ions. Spikes protrude about 8 nm from the capsid surface at the five-fold vertices. The spikes are homopentamers and formed of protein P1. P1 is composed of three beta-barrel domains arranged end to end. The distal C-terminal domains of P1 are used for receptor recognition. The N-termini of P1 form pentagonal assemblies at the vertices. The inner membrane (47 nm in diameter) contains host plasma membrane-derived phospholipids and virus-encoded proteins P3 to P10 (Kivelä et al., 2002). Transmembrane proteins P3 and P6 are organized into a lattice on the membrane vesicle surface, on which the outer protein capsid is assembled (Abrescia et al., 2008). Pseudoalteromonas phage Cr39582 has a nontailed viral particle of about 45 nm capsid in diameter resembling phage PM2 (Leigh et al., 2018).
Physicochemical and physical properties
The mass of the PM2 virion is about 4.5×107 Da and is distributed among protein (72%), lipid (14%) and nucleic acid (14%) (Camerini-Otero et al., 1974, Camerini-Otero and Franklin 1975). The buoyant density of PM2 in CsCl is 1.28 g cm−3 and in sucrose 1.26 g cm−3, and the S20,w is 293 S (Camerini-Otero and Franklin 1975, Kivelä et al., 1999). The Cr39582 virion has a buoyant density of 1.23 g cm−3 in CsCl (Leigh et al., 2018). PM2 virions are stable at pH 6–8, and are very sensitive to ether, chloroform and detergents (Espejo and Canelo 1968). The stablility of PM2 virions is strongly dependent on sodium and calcium ions and virions are sensitive to freezing (Kivelä et al., 2002).
Nucleic acid
The Pseudoalteromonas virus PM2 genome (Männistö et al., 1999) (AF155037) is a highly supercoiled, circular double-stranded (ds) DNA of 10 079 bp (6.6×106 Da). DNA comprises approximately 14% of the virion weight and the G+C content is 42.2%. The genome of Pseudoalteromonas phage Cr39582 is a circular dsDNA of 10 584 bp (MG966533) with a G+C content of 48.2% (Leigh et al., 2018).
Proteins
The Pseudoalteromonas virus PM2 genome has 21 putative genes, 17 of which have been shown to code for structural proteins (P1–P10) and nonstructural proteins (P12–P18; Table 1). Proteins form about 72% of the virus particle weight. The Pseudoalteromonas phage Cr39582 genome has 20 predicted genes (Leigh et al., 2018).
Table 2 Corticoviridae. Pseudoalteromonas virus PM2 proteins.
| Proteina | Mass (kDa) | Location/functionb |
|---|---|---|
| P1 | 37.5 | Spike protein, receptor binding (S) |
| P2 | 30.2 | Major capsid protein (S) |
| P3 | 10.8 | Major membrane protein (S) |
| P4 | 4.4 | Membrane (S) |
| P5 | 17.9 | Membrane (S) |
| P6 | 14.3 | Major membrane protein (S) |
| P7 | 3.6 | Membrane (S) |
| P8 | 7.3 | Membrane (S) |
| P9 | 24.7 | Putative packaging ATPase (S) |
| P10 | 29.0 | Membrane (S) |
| P12 | 73.4 | Replication initiation protein (N) |
| P13 | 7.2 | Transcription factor (N) |
| P14 | 11.0 | Transcription factor (N) |
| P15 | 18.1 | Regulative function (N) |
| P16 | 10.3 | Regulative function (N) |
| P17 | 6.0 | Lysis factor (N) |
| P18 | 5.7 | Lysis factor (N) |
a P is for protein; Arabic numeral corresponds to the Roman numeral of the gene.
b S is for structural protein; N is for non-structural protein.
Lipids
PM2 particles are about 14% lipid by weight (Camerini-Otero and Franklin 1975). The membrane contains 34% phosphatidyl ethanolamine and 66% phosphatidyl glycerol and trace amounts of phosphatidic acid and acyl-phosphatidyl glycerol (Braunstein and Franklin 1971, Camerini-Otero and Franklin 1972, Tsukagoshi et al., 1976). The lipids are derived from the host plasma membrane, but their composition deviates from that of the host bacterium. Lipids form an internal membrane with virus-specific membrane-associated proteins.
Genome organization and replication
To infect and replicate, Pseudoalteromonas virus PM2 delivers its genome across the cell envelope of two known marine host strains: gram-negative Pseudoalteromonas species ER72M2 and BAL-31 (Kivelä et al., 1999). PM2 virions adsorb via the distal tips of the spike proteins to uncharacterized receptors (Abrescia et al., 2008). The internal membrane mediates the translocation of the supercoiled genome across the host cell envelopes, most probably via fusion in a process that is not fully understood (Kivelä et al., 2004). Replication of the viral genome, most probably by a rolling circle mechanism, takes place in proximity to the host cytoplasmic membrane. The largest Pseudoalteromonas virus PM2 gene XII encoding protein P12 shares significant sequence similarity with the superfamily I group of replication initiation proteins (Männistö et al., 1999). The PM2 genome is organized in three operons (Figure 2 Corticoviridae). Operons OEL and OER encode early gene products: the replication initiation protein P12 and transcription regulatory proteins P13, P14, P15 and P16. Expression of the genes for structural proteins is under the control of the late promoter (OL), which is activated by the virus-encoded transcription factors P13 and P14 (Männistö et al., 2003). Mature PM2 virions are released from the cell by lysis. Lysis factor P17 permeabilizes the cytoplasmic membrane and acts like a holin, whereas lysis factor P18 disrupt the outer membrane, and peptidoglycan is most probably disrupted by host lytic factors (Krupovic et al., 2007).
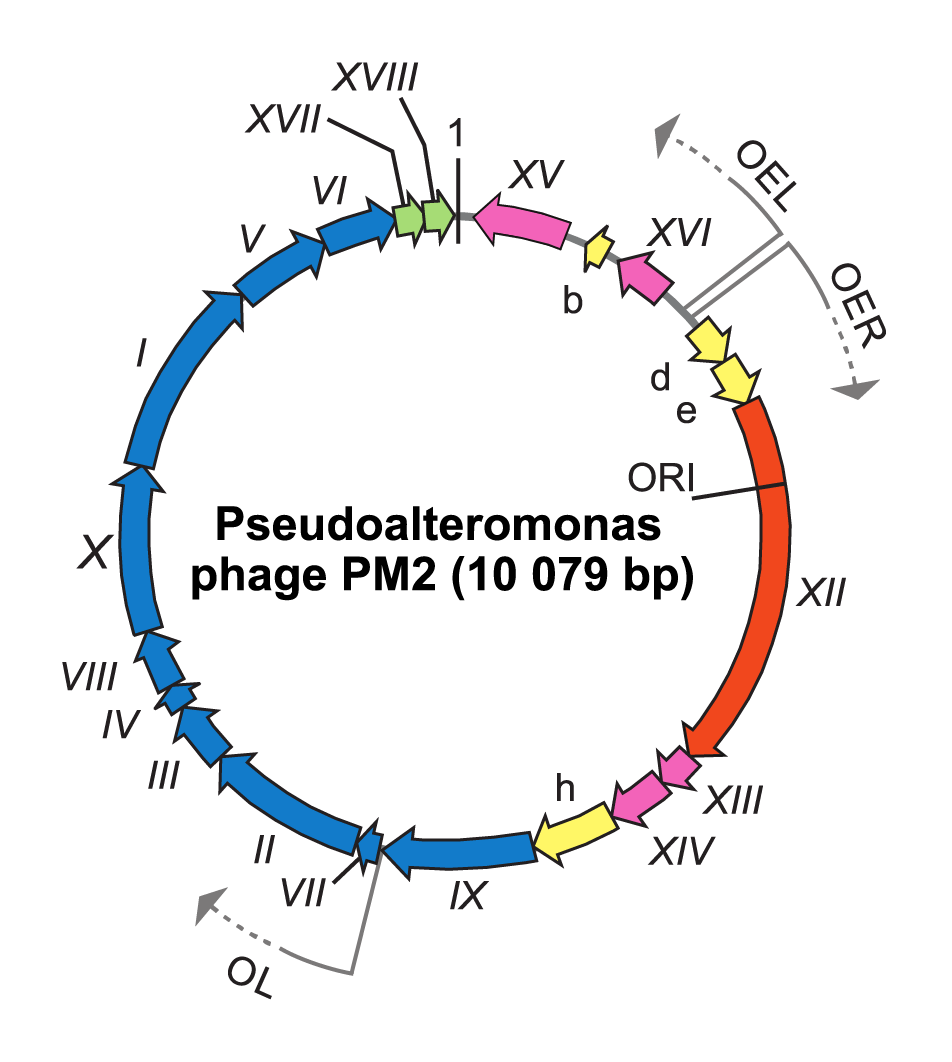 |
| Figure 2 Corticoviridae. Genome organization of Pseudoalteromonas virus PM2. The genome is a 10 079 bp highly supercoiled, circular double-stranded DNA molecule containing 21 genes (Roman numerals) or ORFs (letters) the arrows indicating orientations and three operons (OER, OEL and OL). ORFs known to code for functional proteins are classified as genes and given a Roman numeral. The different colours indicate the ORFs encoding putative proteins (yellow), a gene for replication initiation protein (orange) and the following groups of genes: transcriptional regulation (magenta), structural proteins (blue) and lysis (green). Positions of the origin of replication (ORI) and the first nucleotide (marked as 1) are indicated. |
Biology
Pseudoalteromonas virus PM2 is virulent and replicates in two known strains of marine host bacteria of the genus Pseudoalteromonas. Although the virus is virulent and one of only two members of the family Corticoviridae, comparative genomic approaches have shown that integrated corticoviral genetic elements commonly reside within aquatic bacterial chromosomes (Krupovic and Bamford 2007). Pseudoalteromonas phage Cr39582 is a prophage of Pseudoalteromonas sp. strain Cr6751 from which it can be induced by mitomycin C (Leigh et al., 2018).
Derivation of names
Corticoviridae: from Latin cortex, “crust”, “bark”.
Relationships within the family
The PM2 and Cr39582 genomes are syntenous. There is sequence similarity between them throughout the genome, except for one gene, which corresponds to PM2 gene I, which encodes a spike protein that recognises the host receptor. Full-genome alignment of phages Cr39582 and PM2 shows about 85% overall identity.
Relationships with other taxa
The virion of corticoviruses resembles that of other tailless icosahedral viruses with an internal membrane, such as viruses in the families Tectiviridae, Turriviridae, Simuloviridae, Sphaerolipoviridae, Finnlakeviridae and Matsushitaviridae[OHM1] , which have a lipid bilayer underneath the isometric protein capsid. Pseudoalteromonas virus PM2 major capsid protein is a trimeric protein with two beta-barrels forming hexagonal capsomers (Abrescia et al., 2008). The same viral jelly-roll fold has also been described at least for Salmonella virus PRD1 (family Tectiviridae) (Benson et al., 1999), the archaeal Sulfolobus turreted icosahedral virus 1 (STIV; family Turriviridae) (Maaty et al., 2006), human adenovirus (family Adenoviridae) (Rux et al., 2003), and large eukaryotic viruses Paramecium bursaria Chlorella virus 1 (family Phycodnaviridae) (Nandhagopal et al., 2002) and Chilo iridescent virus (family Iridoviridae), Mimivirus-dependent virus Sputnik (family Lavidaviridae) (Zhang et al., 2012), as well as Flavobacterium phage FLiP (family Finnlakeviridae) (Laanto et al., 2017). Structural conservation of the major capsid proteins in these viruses, led to the establishment of the higher taxa realm Varidnaviria (Koonin et al., 2020, Walker et al., 2020), incorporating also the family Corticoviridae.