Family: Aoguangviridae
Yifan Zhou, Yongjie Wang, Mart Krupovic
The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Mart Krupovic (mart.krupovic@pasteur.fr)
Edited by: Mart Krupovic and Stuart G. Siddell
Posted: November 2023
Summary
The family Aoguangviridae includes dsDNA viruses that have been associated with marine archaea of the order Poseidoniales (formerly known as Marine Group II Euryarchaota). All members of this virus family are known through metagenomics. Virions are predicted to consist of an icosahedral capsid and a helical tail, characteristic of members in the class Caudoviricetes. Aoguangviruses have some of the largest genomes among archaeal viruses and encode most of the components of the DNA replication machinery as well as auxiliary functions. The family Aoguangviridae currently includes a single species, Aobingvirus yangshanense, but many, as of yet, unclassified relatives of this virus group, referred to as ‘magroviruses’, have been discovered by metagenomics in globally distributed marine samples.
Table 1. Characteristics of members of the family Aoguangviridae
| Example | Poseidoniales virus YSH_150918 (ON649702), species Aobingvirus yangshanense, genus Aobingvirus |
| Virion | Predicted to have virions characteristic of members of the class Caudoviricetes, with icosahedral capsids and helical tails |
| Genome | Linear, dsDNA genome of 92.2 kbp |
| Replication | Virus encoded DNA replisome, including family B DNA polymerase |
| Translation | Unknown |
| Host range | Marine archaea of the order Poseidoniales |
| Taxonomy | Realm Duplodnaviria, kingdom Heunggongvirae, phylum Uroviricota, class Caudoviricetes, order Magrovirales: 1 genus and 1 species |
Virion
Morphology
Members of the family Aoguangviridae have been discovered through metagenomics, and have yet to be cultured (Nishimura et al., 2017, Philosof et al., 2017, Xu et al., 2023, Zhou et al., 2023). However, based on the conservation of the structural module characteristic of bacterial and archaeal viruses of the class Caudoviricetes (Baquero et al., 2020) and including the HK97-fold major capsid protein, capsid maturation protease, large subunit of the terminase, portal, major tail protein, baseplate, etc. (Figure 1), virions are predicted to consist of an icosahedral capsid and a helical tail. The lack of the gene encoding a tail sheath protein suggests that the tails of aoguangvirids are not contractile.
Nucleic acid
Aoguangvirids have double-stranded DNA genomes of 90–100 kbp and are among the largest described among archaeal viruses. The 92.3 kb genome of Poseidoniales virus YSH_150918 has a G+C content of 31.5% and encodes 129 predicted proteins (Zhou et al., 2023).
Genome organization and replication
The genome of Poseidoniales virus YSH_150918, the only classified member of the family, has been assembled as a circular molecule, suggesting terminal redundancy and/or circular permutation. The genes encoding functions related to genome replication and virion morphogenesis are organized into two clusters which are predicted to be transcribed in opposite directions (Figure 1.Aoguangviridae). A characteristic feature of aoguangvirids, also observed in other, unclassified viruses of the order Magrovirales, is the extensive set of genes required for genome replication and repair (Nishimura et al., 2017, Philosof et al., 2017, Zhou et al., 2023). In particular, Poseidoniales virus YSH_150918 encodes a family B DNA polymerase, archaeo-eukaryotic DNA primase, DNA polymerase sliding clamp, clamp loader, replicative minichromosome maintenance helicase, ATP-dependent DNA ligase, and other proteins (Zhou et al., 2023). Thus, it is likely that aoguangvirids replicate their genomes with little, if any, requirement for host replication factors.
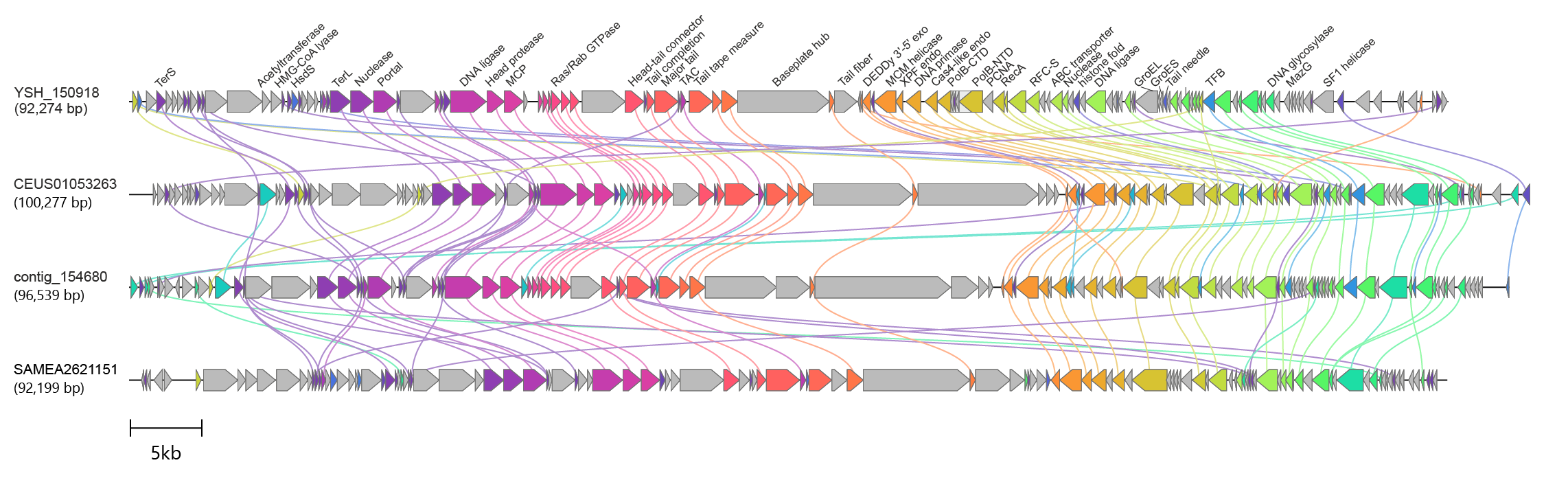 |
| Figure 1.Aoguangviridae. Genome organisation of Poseidoniales virus YSH_150918 and its comparison to the genomes of three unclassified related viruses. Arrows depict open reading frames indicating the direction of transcription. Abbreviations: SF1, superfamily 1; TFB, transcription factor B; RFC-S, replication factor C, small subunit; PCNA, proliferating cell nuclear antigen/DNA polymerase sliding clamp; PolB-NTD/CTD, family B DNA polymerase, N-/C-terminal domain; endo, endonuclease; MCM, minichromosome maintenance; exo, exonuclease; TAC, tail assembly chaperone; MCP, major capsid protein; TerS/TerL, small/large subunit of the terminase; HsdS, type I restriction modification system specificity (S) subunit; HMG-CoA, β-Hydroxy β-methylglutaryl-CoA. |
Biology
No member of the Aoguangviridae has been cultured thus far. Thus, insights into the biology of these viruses can be only gleaned from computation analysis of their genomes. Aoguangvirids do not encode identifiable integrases or transposases and are likely to lead a lytic lifestyle. Besides the genes required for genome replication and repair, as well as virion morphogenesis, aoguangvirids encode auxiliary functions implicated in protein folding (e.g., chaperonin GroEL and cochaperonin GroES) and modulation of the host metabolism (e.g., β-Hydroxy β-methylglutaryl-CoA lyase) (Figure 1.Aoguangviridae).
Derivation of names
Aoguangviridae: after Aoguang, the Dragon King of the East China Sea in ancient Chinese myths and legends, referring to the marine habitat of aoguangvirids and their hosts
Aobingvirus: after Aobing, son of Aoguang
Genus demarcation criteria
The family currently includes a single genus. Sharing more than 60% of proteins has been suggested as a potential genus demarcation criterion; in other words, viruses sharing less than 60% of their proteins would belong to different genera.
Species demarcation criteria
The genus currently includes a single species. Less than 95% overall genome sequence identity has been proposed for species demarcation, that being consistent with the classification of other bacterial and archaeal viruses in the class Caudoviricetes.
Relationships within the family
Aoguangviridae currently includes a single species (Figure 2). However, comparative genomics with sequences of unclassified viruses available in different databases reveals a cluster of related viruses sharing 20–40% of their genes with Poseidoniales virus YSH_150918 (Zhou et al., 2023). These viruses are likely to become members of additional genera in the family Aoguangviridae (Nishimura et al., 2017, Philosof et al., 2017, Xu et al., 2023, Zhou et al., 2023).
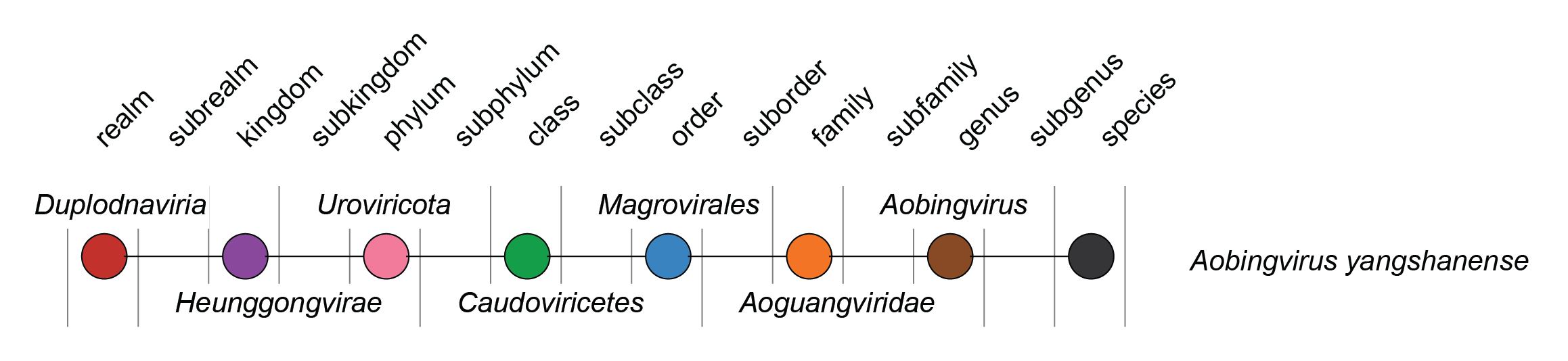 |
| Figure 2. Relationships of the taxa connected to the family Aoguangviridae. |
Relationships with other taxa
Viruses in the family Aoguangviridae are most closely related to unclassified archaeal tailed viruses associated with marine archaea of the order Poseidoniales (Nishimura et al., 2017, Philosof et al., 2017), which based on comparative genomes analysis (Zhou et al., 2023) are likely to form at least two additional families within the order Magrovirales. Collectively, members of the Magrovirales are related to viruses of the family Druskaviridae (class Caudoviricetes) infecting hyperhalophilic archaea (Liu et al., 2021).
Related, unclassified viruses
| Related genomes in GenBank | Accession number |
| TARA_125_MIX_0.22-3_scaffold112370_3 | CEUS01053263* |
* Incomplete genome which can be extended to the total length of 100,277 bp.
Virus names and virus abbreviations, are not official ICTV designations

