Realm: Adnaviria
Mart Krupovic, Fengbin Wang, Virginija Cvirkaite-Krupovic, Jens H. Kuhn, Valerian V. Dolja, David Prangishvili, Edward H. Egelman and Eugene V. Koonin
The citation for this ICTV Report chapter is the summary to be published as Krupovic et al., (2025):
ICTV Virus Taxonomy Profile: Adnaviria 2025, Journal of General Virology, (in press)
Corresponding author: Mart Krupovic (mart.krupovic@pasteur.fr)
Edited by: Evelien Adriaenssens
Posted: March 2025
Summary
Viruses assigned to the realm Adnaviria have linear double-stranded (ds) DNA genomes in which the DNA is structured in the A-form rather than the usual B-form (Baquero et al., 2020b, Wang et al., 2020a). Virions are flexible or rigid filaments, with or without envelopes. Adnavirians share homologous major capsid proteins (MCP) with a distinct α-helical structural fold not observed in other known viruses. The MCPs form homodimeric or heterodimeric capsomers that wrap around and condense the genomic dsDNA into helically symmetric virions. Currently known hosts are hyperthermophilic, thermoacidophilic, and methanotrophic archaea (Table 1 Adnaviria). The realm was established in 2021 to unify the evolutionarily related viruses that share virion structure and organization (Master Species List #36).
Table 1 Adnaviria. Characteristics of viruses of the realm Adnaviria
| Characteristic | Description |
| Example | Sulfolobus islandicus rod-shaped virus 2 (AJ344259), species Icerudivirus hveragerdiense |
| Virion | Flexible or rigid filaments, with or without an envelope, 20–38 nm diameter, 400–2000 nm long, constructed from one or two alpha-helical major capsid proteins |
| Genome | Linear dsDNA of 17.6–41.5 kb, the DNA being in the A-form |
| Replication | Variable |
| Translation | Host translation machinery |
| Host range | Primarily hyperthermophilic and thermoacidophilic archaea of the phylum Thermoproteota |
| Taxonomy | The realm includes 1 kingdom, 1 phylum, 1 class, 3 orders, 6 families, 15 genera, and 33 species |
Virion
Morphology
Virions of adnavirians are filamentous, either without (Rudiviridae) or with (Lipothrixviridae, Ungulaviridae, and Tristromaviridae) an envelope (Figure 1 Adnaviria). Non-enveloped virions are rigid, whereas the enveloped ones are more flexible (Wang et al., 2020a). Virion dimensions are 20–38 nm in diameter and 400–2,000 nm in length. Both ends of the virion are usually capped with diverse terminal structures, which are implicated in host recognition and binding (Pina et al., 2011). Ahmunvirid and chiyouvirid particles have not been experimentally characterized.
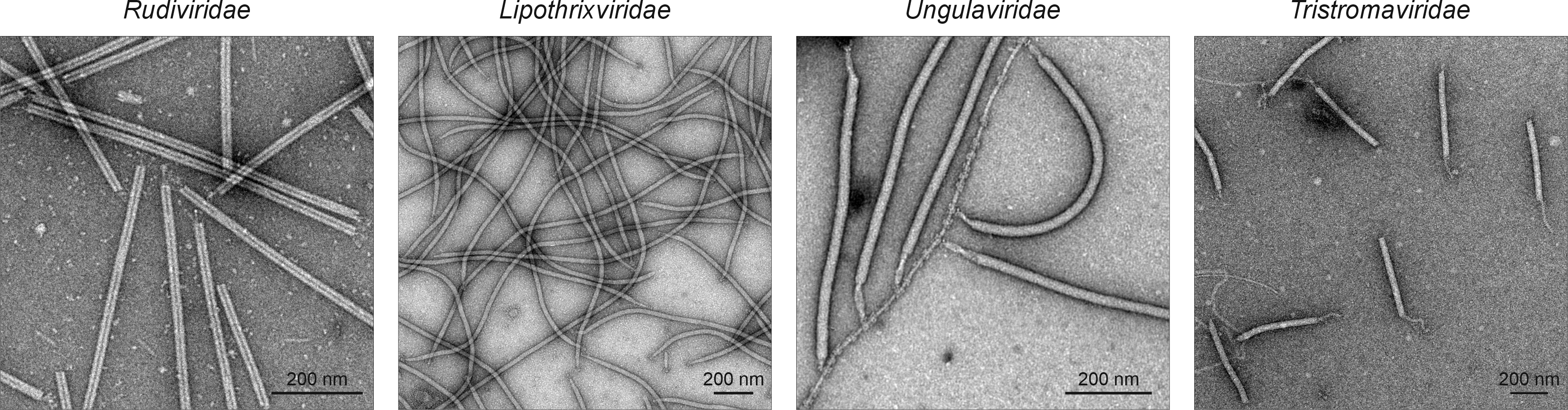 |
| Figure 1 Adnaviria. Electron micrographs of representative adnavirian particles. Particles were negatively stained with 2% (wt/vol) uranyl acetate. Rudiviridae is represented by Saccharolobus solfataricus rod-shaped virus 1, Lipothrixviridae by Sulfolobus islandicus filamentous virus, Ungulaviridae by Acidianus filamentous virus 1, and Tristromaviridae by Pyrobaculum filamentous virus 2. |
Physicochemical and physical properties
The buoyant density in caesium chloride (CsCl) is 1.36 g cm-3 for the non-enveloped virions of rudiviruses and 1.25–1.3 g/cm-3 for enveloped virions. Non-enveloped virions are resistant to treatment with organic solvents and detergents, such as Triton X-100 and TWEEN-20, whereas enveloped virions are sensitive to such treatments (Prangishvili et al., 1999, Rensen et al., 2016, Liu et al., 2018). Solubilization of lipid envelopes exposes nucleocapsid equivalents to the capsids of non-enveloped rudivirids. Both types of virions can be dissociated in the presence of 0.1% sodium dodecyl sulfate (SDS).
Nucleic acid
Adnavirians have genomes comprising linear dsDNA molecules of 17.6–41.5 kb with a guanine–cytosine (GC) content of 24.9–46.6%. As packaged in virions, the genomic DNA is characteristically in the A-form, a dehydrated version of the more typical B-form from which it differs by its more compact helical structure in which base pairs are no longer perpendicular to the helix axis. The partial dehydration of the viral genome results from extensive interactions between the phosphodiester DNA backbone and the major capsid protein molecules.
Proteins
All adnavirians encode homologous major capsid proteins (MCPs) with a unique α-helical fold (Figure 2 Adnaviria). Rudivirids encode a single MCP, whereas members of the other adnavirian families encode two paralogous, structurally similar MCPs (Wang et al., 2020c). High-resolution structures are available for rudivirids (DiMaio et al., 2015, Wang et al., 2020a), lipothrixvirids (Liu et al., 2018, Wang et al., 2020a), ungulavirids (Kasson et al., 2017), and tristromavirids (Wang et al., 2020c). The capsomer, the principal building block of the capsid, is an MCP dimer, a homodimer in rudivirids and a heterodimer in the other adnavirians. The MCP dimers have a closed claw-like organization and tightly wrap around the genomic dsDNA (Figure 2 Adnaviria). Extensive protein–protein and protein–DNA interactions result in coiling and compaction of the viral DNA into helically symmetric virions (Figure 2 Adnaviria). The terminal structures capping the filamentous virions on both ends implicated in host recognition, are composed of minor structural proteins, and usually genus-specific. The genes encoding the major capsid proteins are the only core genes shared by all adnavirians.
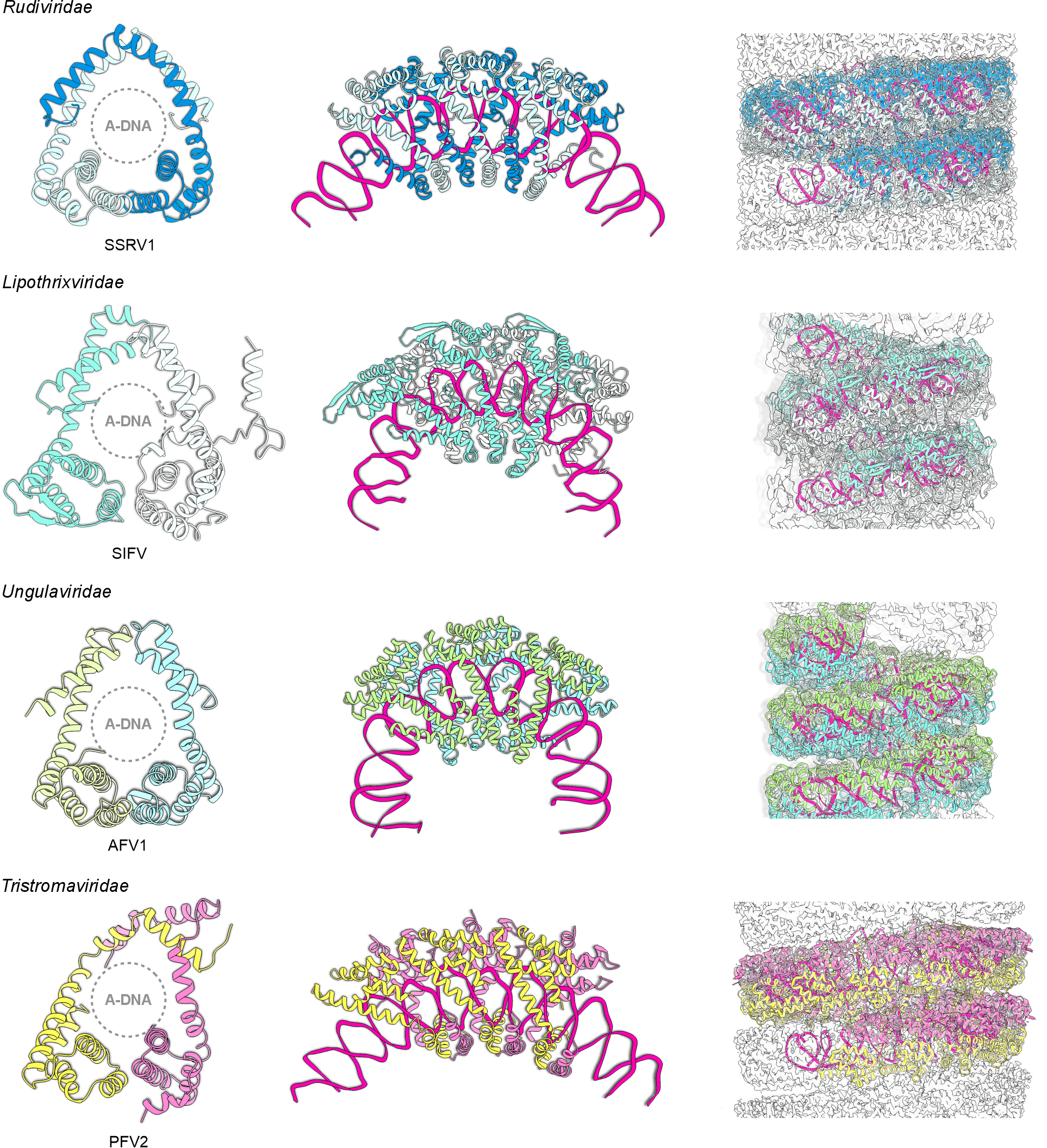 |
| Figure 2 Adnaviria. Major capsid proteins and virion organization of selected adnavirians. Left: The major capsid protein homodimers (in rudivirids) or heterodimers (in all other adnavirians) encircle the A-form dsDNA (A-DNA). Two major capsid protein subunits are shown with different colors. Middle: The major capsid proteins tightly wrap around the genomic DNA. The DNA is shown in magenta. Right: Protein-protein and protein-DNA interactions lead to the coiling of the nucleoprotein filament into a helically symmetrical virion. A segment of the virion is shown in each case. SSRV1, Saccharolobus solfataricus rod-shaped virus 1 (Rudiviridae) (Wang et al., 2020a); SIFV, Sulfolobus islandicus filamentous virus (Lipothrixviridae) (Wang et al., 2020a); AFV1, Acidianus filamentous virus 1 (Ungulaviridae) (Kasson et al., 2017); PFV2, Pyrobaculum filamentous virus 2 (Tristromaviridae) (Wang et al., 2020c). |
Lipids
Virions of enveloped adnavirians, namely, lipothrixvirids, ungulavirids, and tristromavirids, contain host-derived diether and/or tetraether lipids. The envelope surrounding the nucleocapsid of tristromavirids exclusively contains tetraether lipids, with the composition closely matching that of the host membrane (Rensen et al., 2016). In ungulavirids and lipothrixvirids, the lipid envelope is twice as thin as the host cytoplasmic membrane and the ratios of different lipid species in the envelopes are radically different compared to those of the cytoplasmic membranes of the respective hosts (Kasson et al., 2017, Liu et al., 2018, Baquero et al., 2021). In the case of the lipothrixvirid Sulfolobus islandicus filamentous virus, envelopment has been shown to take place de novo in the host cytoplasm by an unknown mechanism (Baquero et al., 2021).
Carbohydrates
Major and minor capsid proteins of adnavirians from the families Rudiviridae, Lipothrixviridae, and Tristromaviridae are glycosylated but the exact structure of the glycans is unknown (Rensen et al., 2016, Liu et al., 2018).
Genome organization and replication
Genomes vary in length and gene arrangement (Figure 3 Adnaviridae).
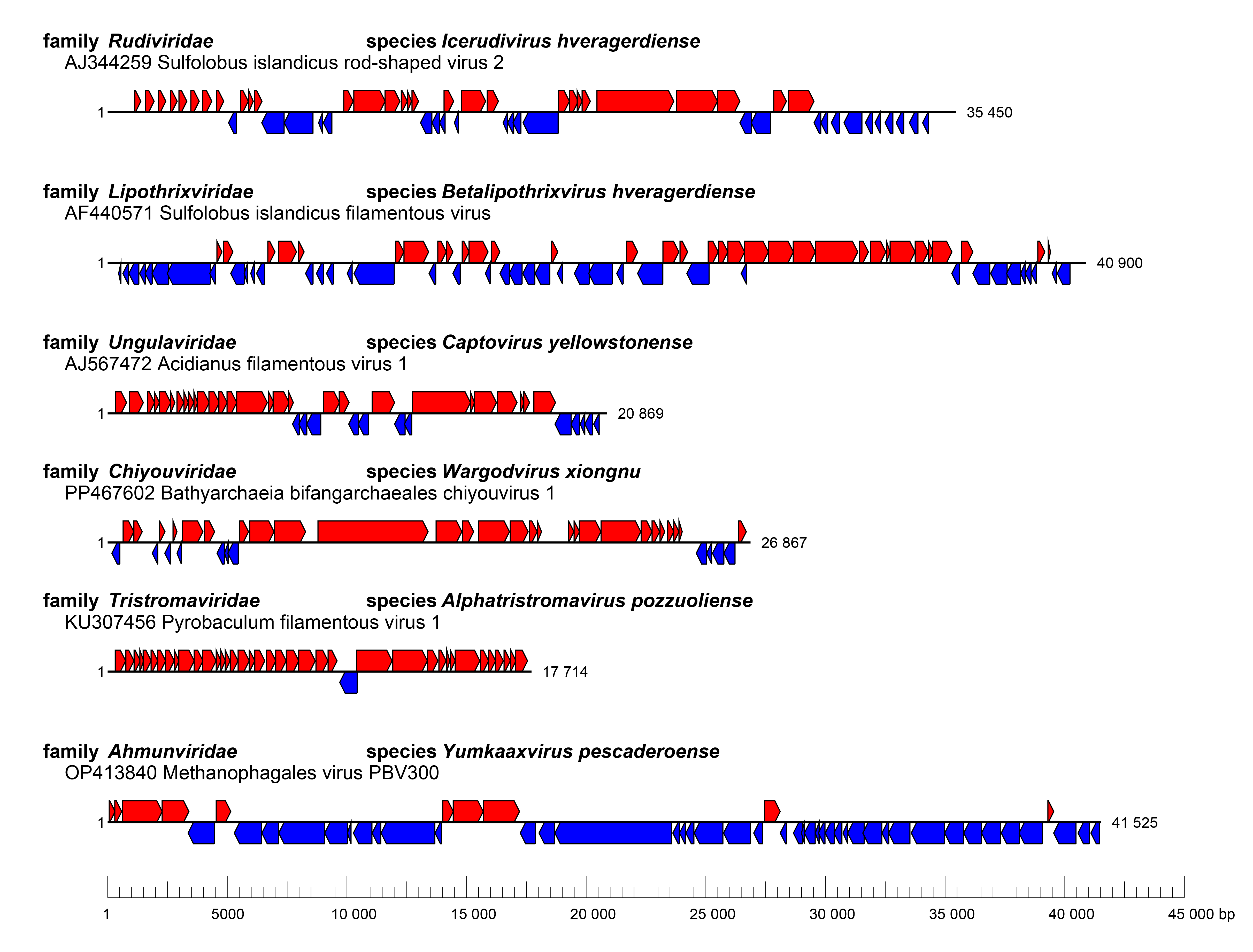 |
| Figure 3 Adnaviria. Genome organization of selected adnavirians. |
Adnavirian genomes are linear and contain terminal inverted repeats. Subterminal regions of some adnavirians, such as ungulavirids, can contain short direct repeats, resembling eukaryotic telomeres. In rudivirids, the two strands of the dsDNA genome are covalently linked, producing hairpins (Blum et al., 2001). The genome of the ungulavirid Acidianus filamentous virus 1 contains a covalently linked terminal protein (Pina et al., 2014).
All rudivirids encode a distinct rolling-circle replication endonuclease of the HUH superfamily which is predicted to initiate genome replication by nicking the dsDNA close to the terminal hairpin (Oke et al., 2011). Genome replication of the rudivirid Sulfolobus islandicus rod-shaped virus 2 (species Icerudivirus hveragerdiense) is mediated through a combination of strand-displacement, rolling-circle, and strand-coupled genome replication mechanisms, which generate multimeric, highly branched, “brush-like” intermediates reaching lengths of >1,200 kb (about 34 genome units) (Prangishvili et al., 2013, Martínez-Alvarez et al., 2016). These intermediates are then processed into unit-length linear genomes with covalently linked hairpin ends.
Replication of Acidianus filamentous virus 1 (family Ungulaviridae, species Captovirus yellowstonense) starts with the formation of a D loop and then progress by the strand-displacement replication mechanism. Termination relies on recombination through the formation of terminal loop-like structures (Pina et al., 2014).
No currently classified adnavirian encodes its own processive DNA polymerase and thus genome replication is likely to be performed by the host replisome. Indeed, the replicative DNA polymerase PolB1 and sliding clamp (PCNA) of the host have been implicated in the replication of rudivirid genomes (Gardner et al., 2014, Martínez-Alvarez et al., 2018). Genome replication of other adnavirians has not been studied experimentally. However, ahmunvirids encode homologs of the archaeo-eukaryotic primase and DNA polymerase sliding clamp that are likely to play key roles in genome replication (Laso-Pérez et al., 2023). Furthermore, partial genomes of putative adnavirians encoding protein-primed family B DNA polymerases have been identified by metagenomics (Tamarit et al., 2022), suggesting that the variety of genome replication strategies used by adnavirians is greater than currently recognized.
Biology
Currently known and predicted hosts of adnavirians are thermophilic archaea of the phylum Thermoproteota, specifically of the orders Sulfolobales (rudivirids, lipothrixvirids, ungulavirids), Thermoproteales (tristromavirids), and Candidatus Bathyarchaeales (chiyouvirids), and anaerobic methanotrophic archaea of the class Candidatus Syntropharchaeia in phylum Halobacteriota (ahmunivirids). Adnavirians are globally distributed, with some groups, namely rudivirids, showing geographical structuring of virus communities in extreme geothermal environments (Baquero et al., 2020a). The association between adnavirians and archaea is predicted to date back to the last archaeal common ancestor (Krupovic et al., 2020).
Interactions between adnavirians and their hosts remain poorly understood, with rudivirids being more extensively studied in this respect. Rudivirids, tristromavirids, and ungulavirids recognize their respective hosts by binding to extracellular filaments, which, in the case of rudivirids and tristromavirids, have been identified as type IV pili (Quemin et al., 2013, Wang et al., 2019, Rowland et al., 2020, Wang et al., 2020b). Upon reaching the cell surface, rudivirid virions disassemble, presumably concomitantly with DNA delivery into the host cytoplasm (Quemin et al., 2013). Once inside the cell, transcription of viral genes commences with the expression of diverse host takeover and counter-defense proteins, such as anti-CRISPR proteins that block the CRISPR defense systems (Quax et al., 2013, Athukoralage et al., 2020, Peng et al., 2020). Notably, Bathyarchaeia bifangarchaeales chiyouvirus 1 (species Wargodvirus xiongnu, family Chiyouviridae) carries two CRISPR mini-arrays, each containing four repeats and three spacers, with the repeats in mini-array 1 being identical to the repeats in the host CRISPR array (Duan et al., 2024). In addition, rudivirids, lipothrixvirids, and ungulavirids inhibit cell division by expressing a homolog of the transcriptional repressor aCcr1 (Li et al., 2023, Yang et al., 2023). Like other known archaeal viruses, adnavirians do not encode their own RNA polymerases and thus rely on the host transcription machinery. However, they encode multiple transcription regulators containing ribbon–helix–helix, winged helix–turn–helix and zinc finger domains, some of which have been experimentally characterized (Guillière et al., 2009, Guillière et al., 2013, Peixeiro et al., 2013). In addition, certain viral genes are under control of host transcription factors (Kessler et al., 2006). Certain rudivirids encode a homolog of the basal transcription factor B, a homolog of the eukaryotic TFIIB (Baquero et al., 2020a), but its role in virus transcription or in virus–host interaction remains uncharacterized.
Virion assembly and envelopment take place in the cytoplasm (Baquero et al., 2021). All characterized adnavirians are lytic viruses, but the mechanisms of lysis seem to vary. Rudivirid and lipothrixvirid particles are released through pyramidal portals, commonly referred to as virus-associated pyramids (VAPs). The VAPs produced by rudivirids are seven-sided (i.e., formed from seven triangular plates) (Bize et al., 2009, Quax et al., 2011), whereas those of lipothrixvirids are six-sided (Baquero et al., 2021). In both cases, VAPs consist of a single small virus-encoded protein (98 aa for the rudivirid SIRV2 and 89 aa for the lipothrixvirid SIFV). VAP formation starts concomitantly with virion assembly. At the end of the infection cycle, when VAPs reach a particular size, the triangular plates come apart like petals of a flower, forming large perforations in the cell envelope through which virions and other cytoplasmic content are released into the extracellular milieu (Daum et al., 2014, Baquero et al., 2021). Tristromavirids appear to rely on a different lysis mechanism, with cells at the end of the infection cycle being slashed open without the discernible presence of defined structures on the cell envelope (Rensen et al., 2016).
Derivation of names
Adnaviria: from A-form DNA characteristic of viruses in this realm; the suffix viria for realm taxa
Zilligvirae: after Wolfram Zillig, a pioneer of research on hyperthermophilic archaeal viruses; the suffix -virae for kingdom taxa
Taleaviricota: from the Latin talea, meaning “rod”, referring to the virion morphology of viruses in the phylum; the suffix -viricota for phylum taxa
Tokiviricetes: from the Georgian თოკი (toki), meaning “thread”, referring to the virion morphology of viruses in the class; the suffix viricetes for class taxa|
Ligamenvirales: from the Latin ligamen, meaning “string, thread”, referring to the virion morphology of viruses in the order; the suffix -virales for order taxa
Maximonvirales: from Maximon, a god of travelers, merchants, medicine men/women, mischief, and fertility in Mayan mythology; the suffix -virales for order taxa; connection unclear
Primavirales: from the Latin prima, meaning “first”, referencing the fact that Thermoproteus tenax virus 1, which is classified in this order, was the first hyperthermophilic archaeal virus to be isolated (in 1983); the suffix -virales for order taxa
Relationships within the realm
Four families are grouped within the order Ligamenvirales, whereas the other two families included in the realm are each allocated to their own order (Figure 4 Adnaviria).
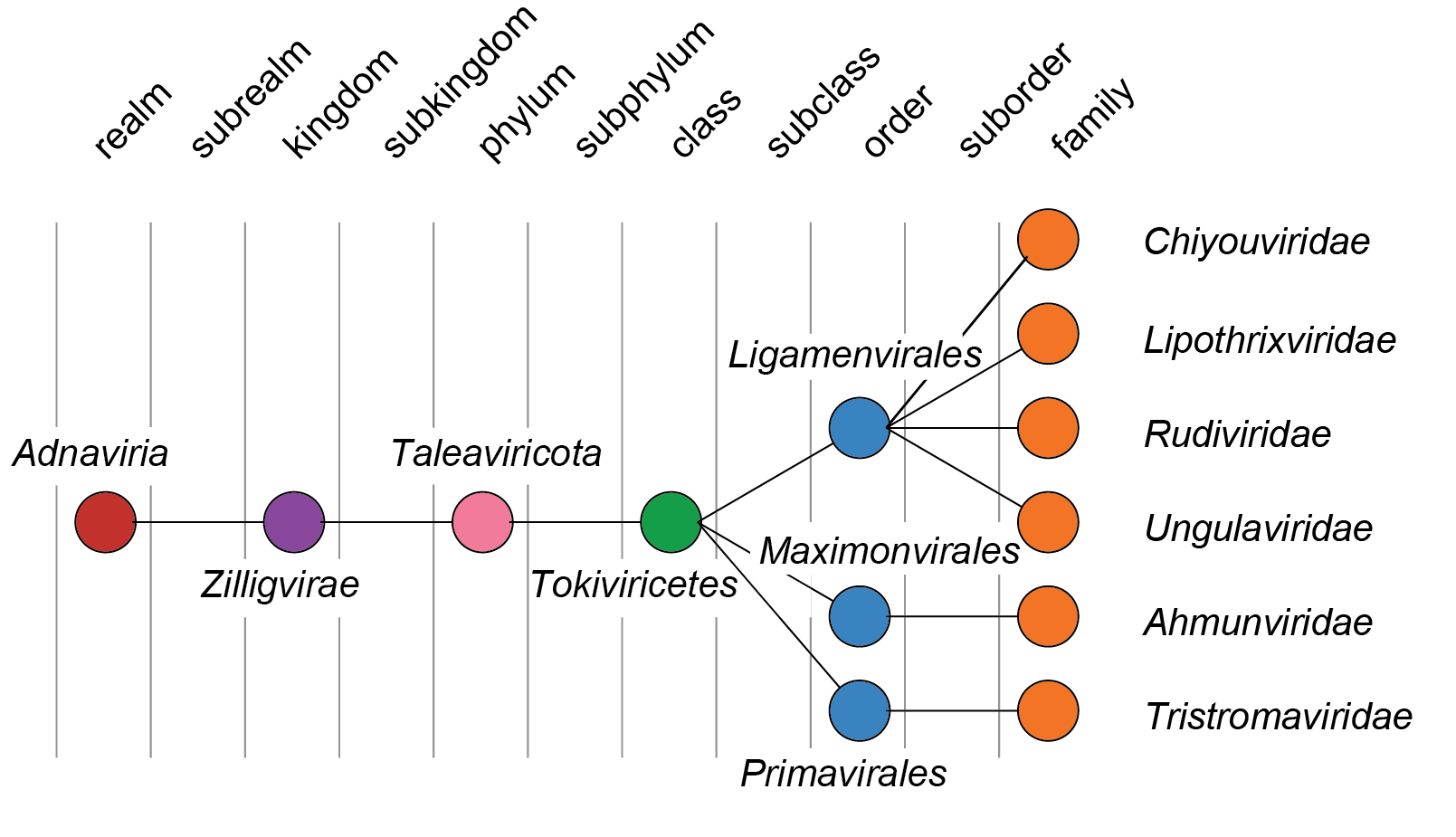 |
| Figure 4 Adnaviria. Taxonomic structure of realm Adnaviria. |

