Family: Globuloviridae
David Prangishvili and Mart Krupovic
The citation for this ICTV Report chapter is the summary published as Prangishvili et al., (2018):
ICTV Virus Taxonomy Profile: Globuloviridae, Journal of General Virology, 99: 1357–1358.
Corresponding authors: David Prangishvili (david.prangishvili@pasteur.fr) and Mart Krupovic (krupovic@pasteur.fr)
Edited by: Andrew M. Kropinski and Stuart G. Siddell
Posted: July 2018, updated March 2025
PDF: ICTV_Globuloviridae.pdf
Summary
The Globuloviridae is a family of enveloped viruses with linear, double-stranded DNA genomes of about 21–28 kbp (Table 1 Globuloviridae). The virions are spherical with a diameter of 70–100 nm and carry a lipid-containing envelope and helical nucleoprotein core. No information is available about genome replication. Globuloviruses infect members of the hyperthermophilic archaea belonging to genera Pyrobaculum and Thermoproteus, which thrive in extreme geothermal environments. Infection does not cause lysis of host cells. The viral genome does not integrate into the host chromosome.
Table 1 Globuloviridae. Characteristics of members of the family Globuloviridae.
| Characteristic | Description |
| Typical member | Pyrobaculum spherical virus (AJ635161), species Alphaglobulovirus obsidianense |
| Virion | Spherical, enveloped with a diameter of 70–100 nm |
| Genome | Linear, dsDNA genomes of about 21–28 kbp |
| Replication | Non-lytic, chronic infection |
| Translation | No information |
| Host range | Hyperthermophilic archaea from the genera Pyrobaculum and Thermoproteus |
| Taxonomy | One genus, four species |
Virion
Morphology
Virions are spherical, 70–100 nm in diameter, with spherical protrusions that are about 15 nm in diameter (Figure 1 Globuloviridae). Virions carry a lipid-containing envelope that encases a superhelical core, consisting of linear dsDNA and nucleoproteins (Figure 1 Globuloviridae) (Ahn et al., 2006, Häring et al., 2004). The morphotype is unusual for dsDNA viruses (Prangishvili 2015, Prangishvili et al., 2017).
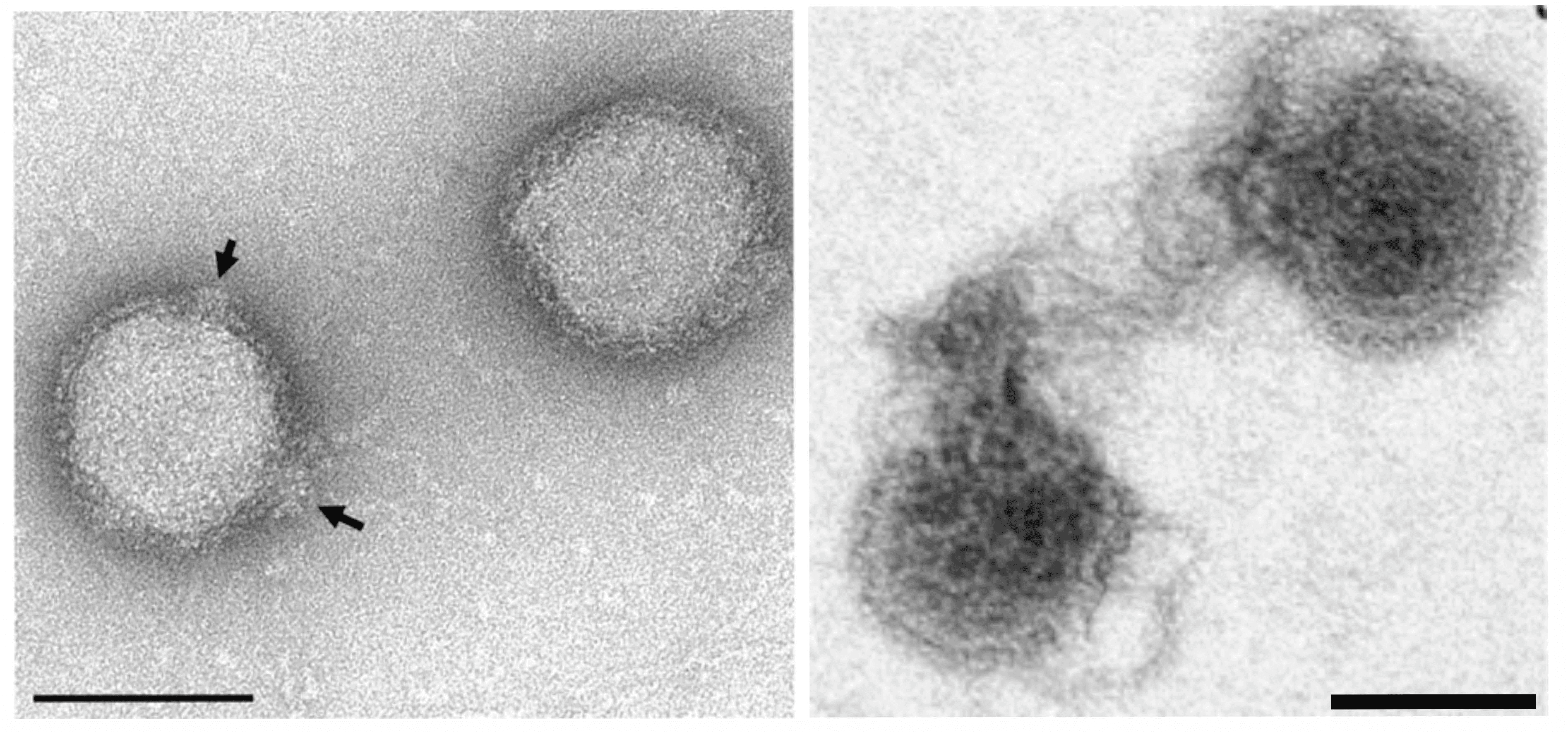 |
| Figure 1 Globuloviridae. Negative-contrast electron micrographs of virions of Pyrobaculum spherical virus. (Left) Intact virions; arrows indicate spherical protrusions. (Right) Partially disrupted virions extruding disordered nucleoprotein core. The bars represent 100 nm. Modified from (Häring et al., 2004). |
Physicochemical and physical properties
Virion buoyant density in sucrose is about 1.3 g cm−3. Virions are highly heat-stable. Prolonged exposure to oxygen does not affect the efficiency of infection.
Nucleic acid
Virions contain a single molecule of linear dsDNA, comprising 28,337 bp for Pyrobaculum spherical virus (PSV).
Proteins
Virions contain three major structural protein with molecular masses of about 16 (VP1), 20 (VP2) and 33 (VP3) kDa (Ahn et al., 2006, Häring et al., 2004). About 80% of amino acids of the major structural protein VP3 are hydrophobic.
Lipids
Virion contains lipids selectively acquired from the pool of host lipids.
Carbohydrates
None reported.
Genome organization and replication
Virions contain a single molecule of linear dsDNA, comprising 28,337 bp for PSV and ~21.6 kbp for Thermoproteus tenax spherical virus 1 (TTSV1). The ends of the linear genome carry inverted repeats (190 bp for PSV), which contain multiple copies of 5 bp direct repeats. In the case of PSV, the two strands of the dsDNA genome appear to be covalently linked at the termini (Häring et al., 2004). PSV and TTSV1 genomes have 48 and 38 open reading frames (ORF), respectively, of which only 15 are shared between the two viruses. Several examples of gene duplication have been reported (Fig. 2). Almost all predicted genes are located on one DNA strand and do not show sequence similarity to genes in extant databases (Krupovic et al., 2018). High resolution structures have been solved for 5 proteins of PSV (Figure 2 Globuloviridae), shedding light on the potential functions of some of these proteins (Krupovic et al., 2012). ORF165a and its paralogs, ORF165b and ORF236, contain a winged helix-turn-helix domain and might have a DNA-binding capacity. Similarly, ORF126 and ORF107 contain a C4-type zinc-binding domain and might be involved in transcription regulation. Finally, ORF137 displays unexpected structural similarity to Sm-like RNA-binding proteins, such as bacterial Hfq, and might be involved in the metabolism of viral or host RNA molecules, as is the case for cellular Sm-like proteins. Globuloviruses do not encode identifiable genome replication proteins and are likely to recruit the host machinery for genome replication.
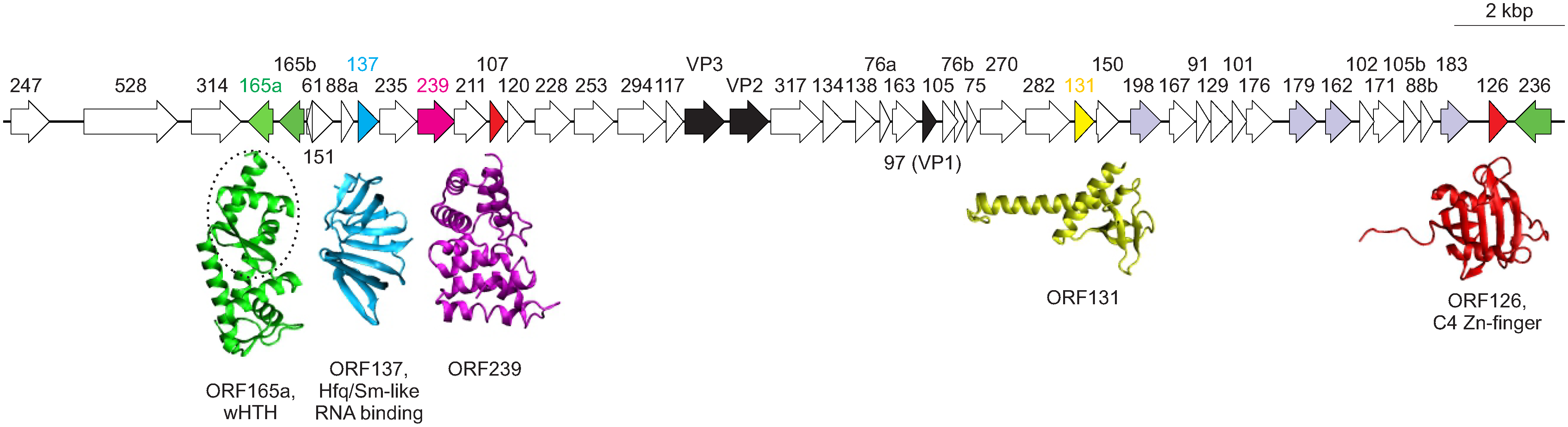 |
| Figure 2 Globuloviridae. Genome organization of Pyrobaculum spherical virus showing location, sizes and direction of putative genes. Genes encoding the three major structural proteins, VP1-3, are shown in black. Paralogous genes are indicated with the same colors. The available protein X-ray structures are shown beneath the genome map and are shaded with distinct colours that follow the shading of their corresponding genes. The winged helix-turn-helix (wHTH) domain of ORF165a is circled (modified from (Krupovic et al., 2012)). |
Biology
PSV infects hyperthermophilic archaea from the genera Pyrobaculum and Thermoproteus (Häring et al., 2004), whereas TTSV1 has been shown to replicate in a single strain of Thermoproteus (Ahn et al., 2006). The hosts are anaerobes thriving in extreme geothermal environments with temperatures around 85 °C, pH 6. Globuloviruses establish a chronic infection and are released from the host cells without causing lysis. The viral genome does not integrate into the host chromosome.
Derivation of names
Globuloviridae: from Latin globulus, “small ball”.
Relationships within the family
Viruses from the two species, Alphaglobulovirus obsidianense and Alphaglobulovirus cinderense, share less than half of their genes (Ahn et al., 2006, Häring et al., 2004). Viruses from the two species in the genus differ in virion size, host range, and in the size and nucleotide sequence of the genome.
Relationships with other taxa
No relationships known.

