Family: Lebotiviridae (Interim Report)
This is a summary page created by the ICTV Report Editors using information from associated Taxonomic Proposals and the Master Species List.
Edited by: Holly Hughes
Posted: October 2024
Summary
The family Lebotiviridae includes dsRNA viruses of invertebrates (Table 1 Lebotiviridae). The family Lebotiviridae was established in 2024 (Master Species List #39). The family consists of one genus, Lebotivirus, and 8 species.
Table 1 Lebotiviridae. Characteristics of members of the family Lebotiviridae.
| Characteristic | Description |
| Example | Leptopilina boulardi toti-like virus (KF274642), species Lebotivirus ichi, genus Lebotivirus |
| Virion | Non-enveloped, icosahedral particles of 40 nm in diameter |
| Genome | A linear, bicistronic dsRNA of 6.7–8.1 kbp (Figure 1 Lebotiviridae) |
| Replication | Unknown |
| Translation | RdRP possibly expressed by −1 ribosomal frameshifting |
| Host range | Invertebrates |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Duplornaviricota, class Chrymotiviricetes, order Ghabrivirales: 1 genus and 8 species (Figure 2 Lebotiviridae) |
Virion
Morphology
Lebotivirids have been shown to have an icosahedral non-enveloped virion about 40nm in diameter (Martinez et al., 2016).
Nucleic acid
The nucleic acid is a single segment of dsRNA (about 6.7–8.1kb), encoding two genes in an overlapping reading frame resulting from a −1 ribosomal frame shift (Martinez et al., 2016).
Proteins
An upstream open reading frame encodes the capsid protein, and the downstream open reading frame encodes the RNA-dependent RNA polymerase (RdRP) following a −1 ribosomal frame shift (Martinez et al., 2016, Viljakainen et al., 2018).
Genome organization and replication
Details of the replication of lebotivirids are unknown. The translation of the two gene products is believed to occur through the production of a fusion protein of the capsid and RdRP that results from a −1 ribosomal frame shift (Martinez et al., 2016, Viljakainen et al., 2018) (Figure 1 Lebotiviridae).
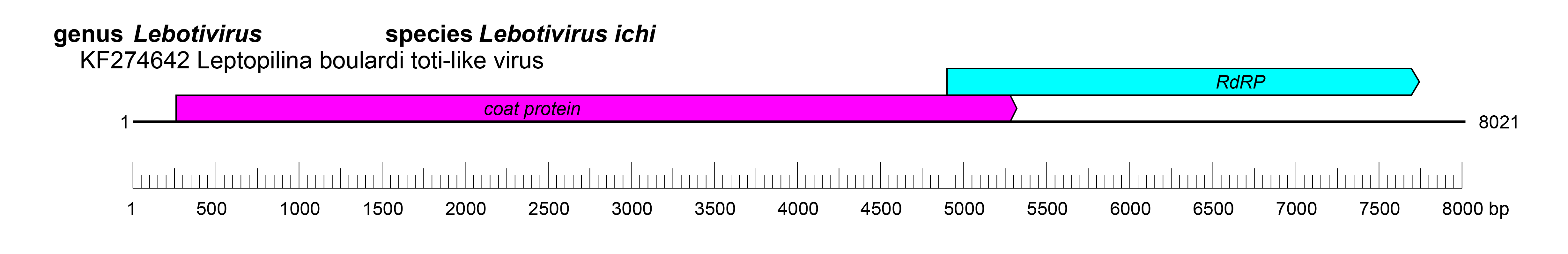 |
| Figure 1 Lebotiviridae. Genome organisation of Leptopilina boulardi toti-like virus, a member of the family Lebotiviridae. Boxes indicate open reading frames as annotated on GenBank accession KF274642. |
Biology
Lebotivirids have been identified in a range of invertebrate insects. Leptoilina boulardi toti-like virus was isolated from a parasitic wasp (Leptopilina boulardi) in which it was shown to be vertically transmitted and potentially provide a fitness advantage to its parasitoid host (Martinez et al., 2016). Additional lebtoviruses have been identified by high-throughput sequencing of ants (Linepithema humile) (Viljakainen et al., 2018), fire ant (Solenopsis invicta) (Valles and Rivers 2019), water strider, horsefly, and pools of insects (Shi et al., 2016) throughout the world.
Derivation of names
Lebotiviridae, Lebotivirus: from Leptopilina boulardi toti-like virus, a member of the family; the suffix viridae for family taxa or the suffix -virus for genus taxa. Epithets are derived from Latinized Japanese numbers (Figure 2 Lebotiviridae).
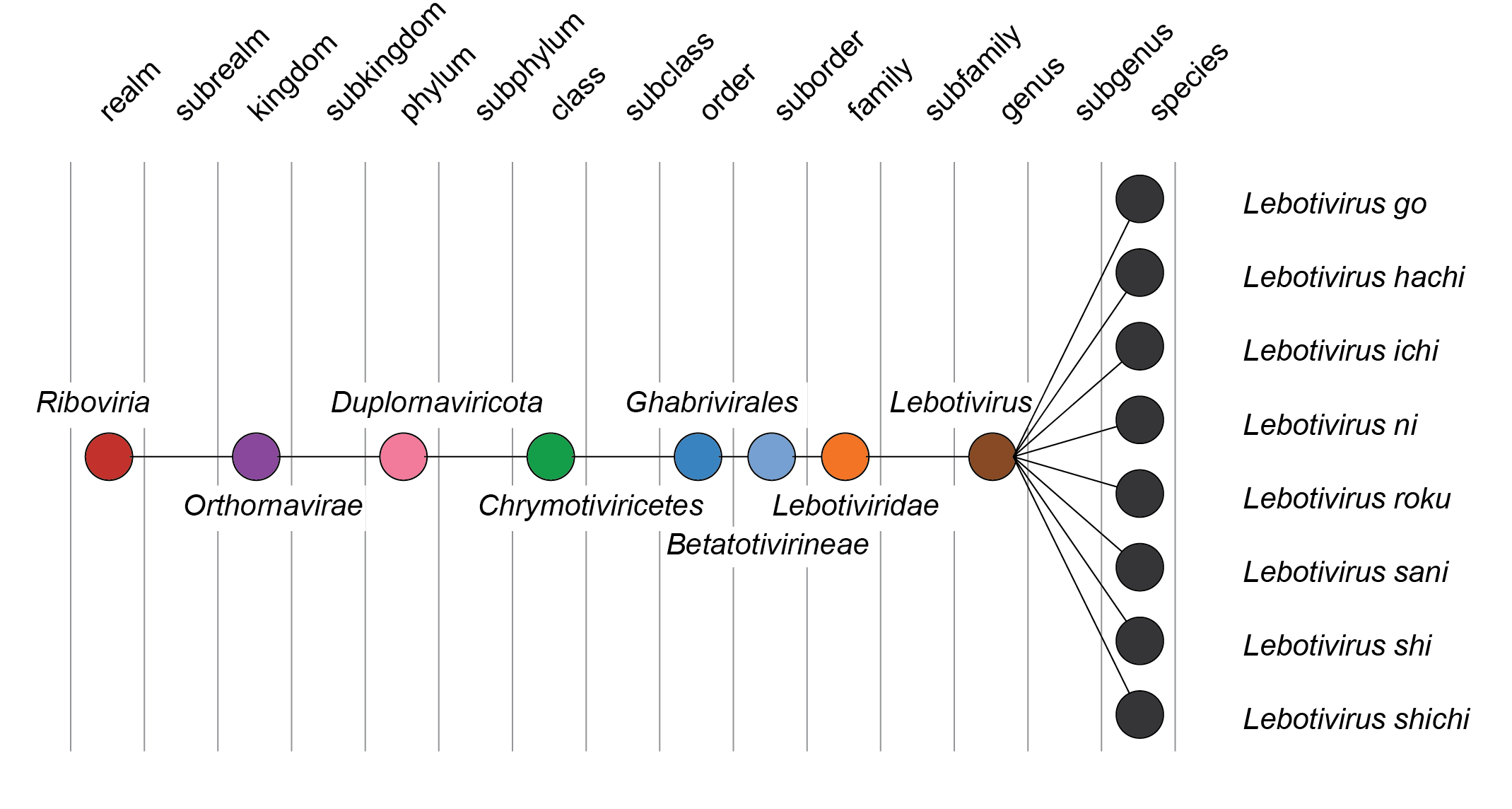 |
| Figure 2 Lebotiviridae. Taxonomy of the family Lebotiviridae. |
Species demarcation criteria
1) The host species for the virus must be assigned convincingly.
2) For viral genome sequences assembled from high-throughput sequencing reads, and not validated in full by Sanger sequencing, read numbers, RPKM values, and/or coverage depth values for the assembled sequence must be specified, or the read data must be available at NCBI such that these values can be determined.
3) Virus genome sequences that share <70% RdRP amino acid sequence identity will be considered to represent different virus species regardless of whether reported from the same or different host species.
4) Virus genome sequences that share ![]() 70% RdRP amino acid sequence identity, but are reported from different host species, are considered to represent the same virus species.
70% RdRP amino acid sequence identity, but are reported from different host species, are considered to represent the same virus species.

