Family: Tulasviridae
Jens H. Kuhn, Scott Adkins, Katherine Brown, Juan Carlos de la Torre, Michele Digiaro, Holly R. Hughes, Sandra Junglen, Amy J. Lambert, Piet Maes, Marco Marklewitz, Gustavo Palacios, Takahide Sasaya (笹谷孝英), Yong-Zhen Zhang (张永振) and Massimo Turina
The citation for this ICTV Report chapter is the summary published as:
Kuhn et al., (2023) ICTV Virus Taxonomy Profile: Tulasviridae 2023, Journal of General Virology 2023, 104, 001933
Corresponding author: Massimo Turina ([email protected])
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: November 2023, updated January 2025
Summary
Tulasviridae is a family of ambisense RNA viruses with genomes of about 12.2 kb (Table 1 Tulasviridae). These viruses have been found in fungi. The family includes two genera with two species. The tulasvirid genome is nonsegmented and contains two or three open reading frames (ORFs) that encode a nucleoprotein (NP), a large (L) protein containing an RNA-directed RNA polymerase (RdRP) domain, and a protein of unknown function (X).
Table 1 Tulasviridae. Characteristics of members of the family Tulasviridae
| Characteristic | Description |
| Example | Tulasnella bunyavirales-like virus 1 (MN793997), species Orthotulasvirus tulasnellae, genus Orthotulasvirus |
| Virion | Unknown |
| Genome | About 12.2 kb of nonsegmented ambisense RNA |
| Replication | Unknown |
| Translation | Unknown |
| Host range | Agaricomycete fungi |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Bunyaviricetes, order Elliovirales; the family includes two genera and two species |
Virion
Morphology
Unknown
Nucleic acid
Tulasvirids have a nonsegmented, linear, ambisense RNA genome with a total length of about 12.2 kb (Sutela et al., 2020).
Genome organization and replication
Viruses of the family Tulasviridae have a nonsegmented ambisense genome with two or three ORFs that encode an NP, an X, and an L protein containing an RdRP domain (Sutela et al., 2020) (Figure 1 Tulasviridae).
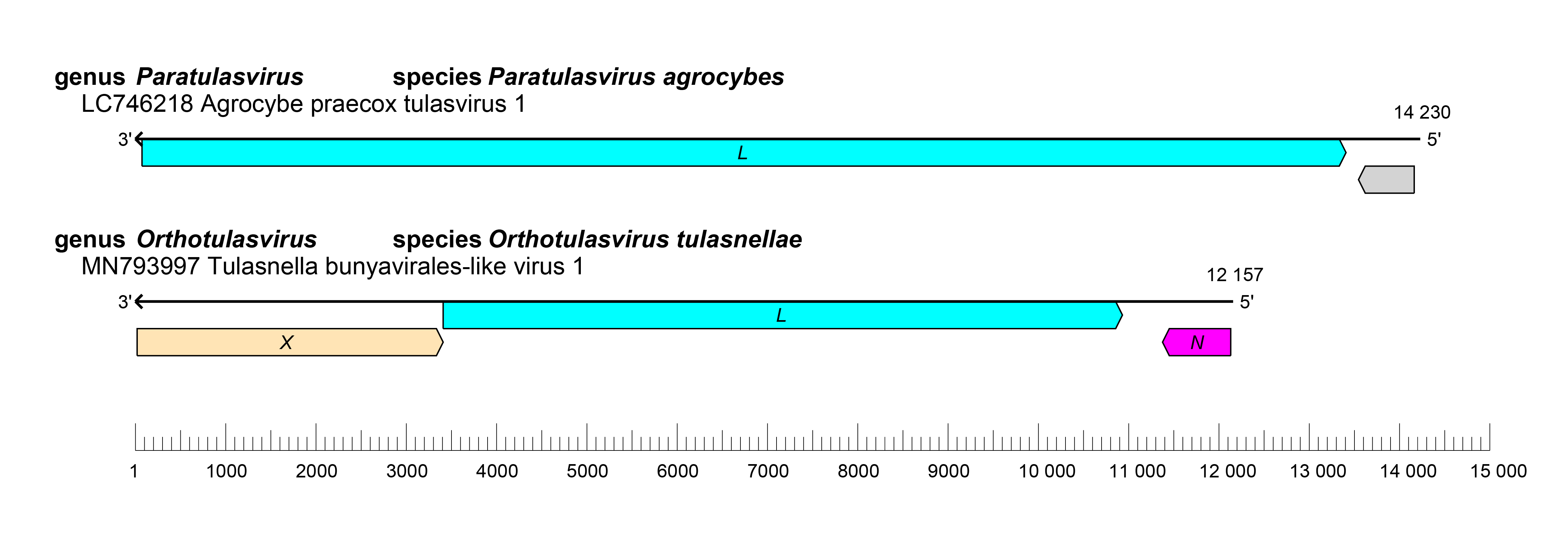 |
| Figure 1 Tulasviridae. Genome organization of Agrocybe praecox tulasvirus 1 and Tulasnella bunyavirales-like virus 1. ORFs are colored according to the predicted protein function (L, large protein gene; NP, nucleoprotein; X, gene encoding a protein of unknown function). |
Biology
Tulasnella bunyavirales-like virus 1 (TB-LV1), was detected in unspecified orchid mycorrhizal agaricomycete fungi (tulasnellaceaen Tulasnella sp.) in association with long-lipped serapias (orchidacean Serapias vomeracea (Burm.f.) Briq.) in Italy (Sutela et al., 2020).
Agrocybe praecox tulasvirus 1 was found in agaricomycete fungi (strophariaceaen Agrocybe praecox (Pers.) Fayod) in Japan (Zhao et al., 2023).
Derivation of names
agrocybes: from the host fungus Agrocybe praecox (Pers.) Fayod
Orthotulasvirus: from the Greek ortho, meaning “straight” and the host genus Tulasnella
Paratulasvirus: from the Greek para, meaning “next to” and the host genus Tulasnella
tulasnellae, Tulasviridae: from the host genus Tulasnella
Genus demarcation criteria
Members of the genus Orthotulasvirus are monophyletic and have a genome with three ORFs. The only member of the genus Paratulasvirus is phylogenetically distinct and its genome has two ORFs.
Relationships within the family
Two distinct genome organizations characterize the members of the two established genera; for both genera the ambisense RNA genome encodes a putative RdRP and a putative N protein.
Relationships with other taxa
Viruses of the family Tulasviridae are most closely related to bunyaviral crulivirids, fimovirids, peribunyavirids, phasmavirids, and tospovirids (Huang et al., 2019, Sutela et al., 2020) (Figure 2 Tulasviridae).
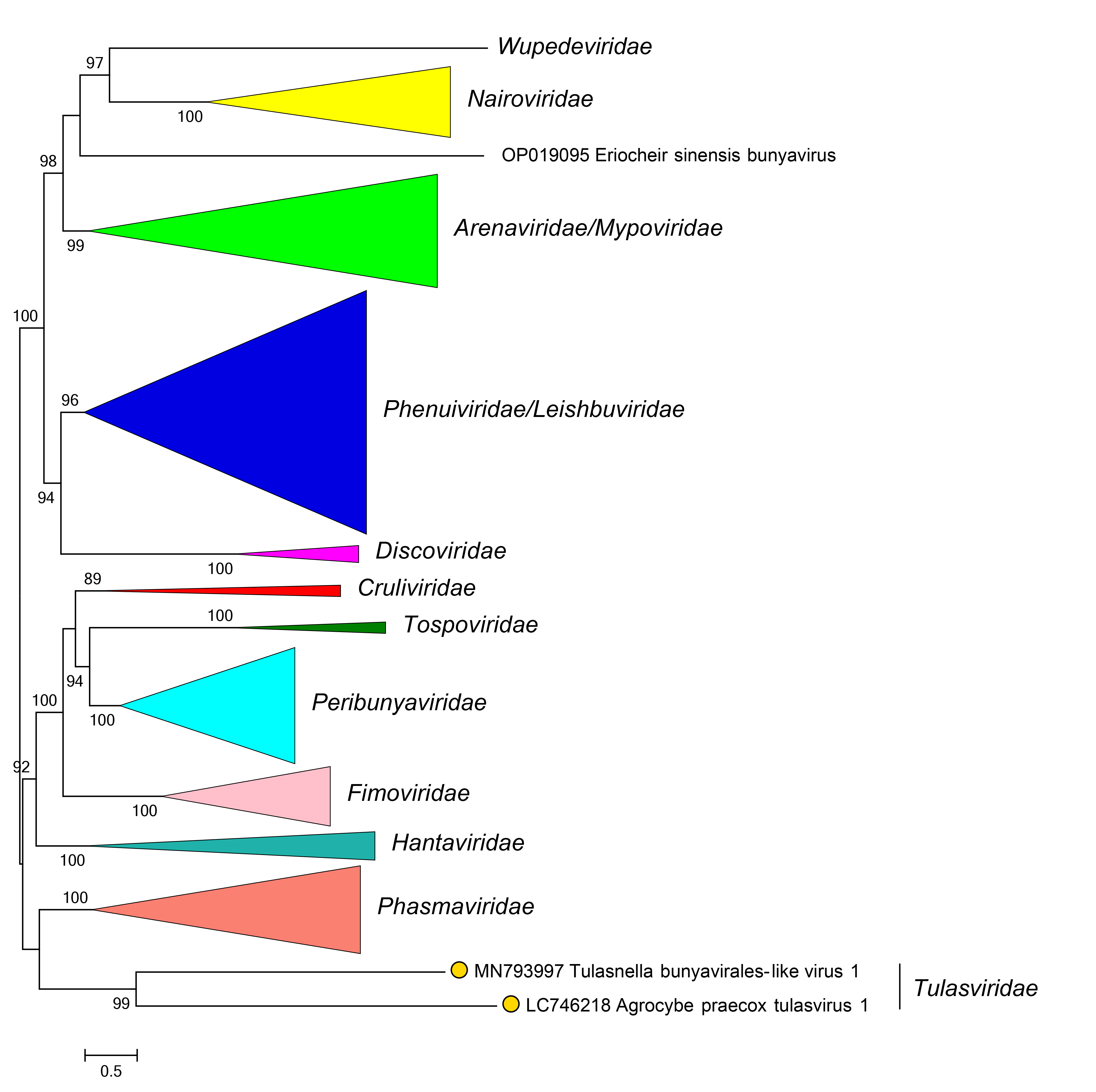 |
| Figure 2 Tulasviridae. Phylogenetic relationships of Tulasnella bunyavirales-like virus 1 with other members of the order Bunyavirales. L protein sequences were aligned using MUSCLE and a maximum likelihood tree was produced using FastTree with default settings. Family branches are collapsed. Numbers at nodes indicate bootstrap support where this was >70%. |
Related, unclassified viruses
| Virus name | Accession number | Reference |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-0928–1 |
| (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-0928–2 | OQ999728 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-0928–3 | OQ999749 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-0928–4 | OQ999775 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-0928–5 | OQ999745 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-0928–6 | OQ999724 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-0928–7 | OQ999764 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-0928–8 | OQ999778 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-0942–1 | OQ999774 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-0942–2 | OQ999769 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-0942–3 | OQ999707 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-1084–1 | OQ999737 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-1084–2 | OQ999734 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-1084–3 | OQ999772 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-1084–4 | OQ999758 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-1084–5 | OQ999731 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-10125–1 | OQ999781 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-10125–2 | OQ999700 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-10125–3 | OQ999736 | (Li et al., 2023) |
| Rhizoctonia cerealis bunyavirus isolate RcBYV-10125–4 | OQ999746 | (Li et al., 2023) |
Virus names and virus abbreviations are not official ICTV designations.

