Family: Paramyxoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are 150 nm or more in diameter, pleomorphic, but usually spherical in shape, although filamentous and other forms are common. Virions consist of a lipid envelope surrounding a nucleocapsid. The envelope is derived directly from the host cell plasma membrane by budding and contains two or three transmembrane glycoproteins. These are present as homo-oligomers and form spike-like projections, 8–12 nm in length, spaced 7–10 nm apart (depending on the genus). One non-glycosylated membrane or matrix protein is associated with the inner face of the envelope. The viral nucleocapsid consists of a single species of viral RNA and associated proteins. It has helical symmetry and is 13–18 nm in diameter with a 5.5–7 nm pitch (depending on the subfamily); its length can be up to 1000 nm in some genera. Multiploid virions are found, although the vast majority of virions contain a single functional genome. The viral polymerase is packaged in the virion.
(Right) Negative contrast electron micrographs of intact parainfluenza virus 5 (PIV5, previously known as simian virus 5 [SV5]) particles (genus Rubulavirus) (top) and the PIV5 nucleocapsid after detergent lysis of virions (bottom) (courtesy of G.P. Leser and R.A. Lamb). The bars represent 100 nm. (Left top and bottom) Schematic diagrams of PIV5 particles in cross-section (N) (formerly NP), nucleocapsid; P, phospho-protein; L, large polymerase protein; V, cysteine rich protein that shares its N-terminus with P sequence and for PIV5 is found in virions; M, matrix or membrane protein; F, fusion protein; HN, hemagglutinin-neuraminidase; SH, small hydrophobic protein). (Adapted from Kingsbury, D.W. (1990). Paramyxoviridae: the viruses and their replication. In: Virology, 2nd edn (B.N. Fields and D.M. Knipe, Eds.), Raven Press, New York; and from Scheid, H. (1987). In: Animal Virus Structure (M.V. Nermut, and A.C. Steven, Eds.), Elsevier, Amsterdam; with permission.)
Physicochemical and physical properties
Virion Mr is about 500 ×106, and much greater for multiploid virions. Virion buoyant density in sucrose is 1.18–1.20 g cm−3. Virion S20,w is at least 1000S. Virions are very sensitive to heat, lipid solvents, ionic and non-ionic detergents, formaldehyde and oxidizing agents.
Nucleic acid
Virions contain a single molecule of linear, negative sense, ssRNA that is not infectious alone but is infectious in the form of the nucleocapsid. The RNA genome size varies substantially: 15,384 nt for Sendai virus (SeV); 15,600 nt for human parainfluenza virus 1 (HPIV1); 15,462 nt for human parainfluenza virus 3, (HPIV-3); 15,384 nt for mumps virus (MuV); 15,246 nt for parainfluenza virus 5 (PIV5, previously known as simian virus 5 [SV-5]); 15,450 nt for simian virus 41 (SV-41); 15,156 nt for Newcastle disease virus (NDV); 14,904–17,262 for other avulaviruses; 15,654 nt for human parainfluenza virus 2 (HPIV-2); 18,234 nt for Hendra virus (HeV); 18,246–15,252 nt for Nipah virus (NiV); 15,894 nt for measles virus (MeV); 15,690 nt for canine distemper virus (CDV); 15,882 nt for rinderpest virus (RPV); 15,702 nt for cetacean morbillivirus (CeMV); 15,191–15,226 nt for human respiratory syncytial virus (HRSV), 13,280–13,378 nt for human metapneumovirus (HMPV) and 19,212 nt for Beilong virus (BeiPV), an unclassified virus in the subfamily Paramyxovirinae. Genome lengths for all viruses in the subfamily Paramyxovirinae are multiples of 6, which is a requirement for the efficient replication of the members of the subfamily Paramyxovirinae, but does not apply to the members of the subfamily Pneumovirinae. Some virions may contain positive sense RNA. Thus, partial self-annealing of extracted RNA may occur. Intracellularly, or in virions, genome-size RNA is found exclusively as nucleocapsids. The genome RNA does not contain a 5′ cap, nor a covalently linked protein. The genome 3′ end is not polyadenylated.
Proteins
Members of the subfamily Paramyxovirinae encode 7–10 proteins (5–250 kDa) of which 2–4 (or more) are derived from the 2–3 overlapping ORFs in the P locus (Figure 2). Pneumoviruses encode 9–11 proteins of 4.8–250 kDa, including two proteins encoded by overlapping ORFs in the M2 locus. Virion proteins common to all genera include: three nucleocapsid-associated proteins, i.e., an RNA-binding protein (N) (formerly NP), a phosphoprotein (P) and a large polymerase protein (L); three membrane-associated proteins, i.e., an unglycosylated inner membrane or matrix protein (M); and two glycosylated envelope proteins, comprising a fusion protein (F) and an attachment protein (G, or H, or HN). The F protein is synthesized within an infected cell as a precursor (F0) that is activated following cleavage by cellular protease(s) to produce the virion disulfide-linked F1 and F2 subunits (order: amino F2-S-S-F1 carboxyl). Variable proteins include putative non-structural proteins (C, NS1, NS2), a cysteine-rich protein that binds zinc (V) (in the subfamily Paramyxovirinae only) that can be structural or non-structural depending on the virus, a small integral membrane protein (SH), a transcription processivity factor (M2-1, formerly called 22K protein) which previously was thought to be a second M-like protein, and a non-abundant protein (M2-2) involved in the balance between genome replication and transcription. Virion enzyme activities (variously represented among the genera) include an RNA-dependent RNA transcriptase, mRNA guanylyl and methyl transferases, and a neuraminidase. A protein kinase is associated with many members but it is probably of cellular origin.
Lipids
Lipids in the viral envelope are derived from host cell plasma membrane.
Carbohydrates
Virions are composed of 6% carbohydrate by weight; composition is dependent on the host cell. Fusion and attachment proteins are glycosylated by N-linked carbohydrate side chains. In the subfamily Pneumovirinae the attachment protein (G) is heavily glycosylated by O-linked as well as N-linked carbohydrate side chains. The SH protein of respiratory syncytial virus contains polylactosaminoglycan.
Genome organization and replication
The genome organization is illustrated in Figure 2 for viruses representing the seven genera of the family. After attachment to cell receptors, virus entry is achieved by fusion of the virus envelope with the cell surface membrane. This can occur at neutral pH. Virus replication occurs in the cell cytoplasm and is thought to be independent of host nuclear functions. The genome is transcribed processively from the 3′ end by virion-associated enzymes into 6–10 separate, subgenomic, positive sense mRNAs. Transcription is guided by short (10–13 nt) conserved transcription start and termination/polyadenylation signals flanking each transcriptional element. The mRNAs are capped and possess 3′-poly(A) tracts synthesized by reiterative copying of the polyadenylation site. Intergenic regions are either highly conserved in sequence and length (Respirovirus, Henipavirus, Morbillivirus and all of the newly discovered viruses in the unassigned group; Figure 2 and list on p. 683 below) or are not conserved in sequence and length (Rubulavirus, Avulavirus, Pneumovirinae). RNA replication occurs through an intermediate, the antigenome, that is a complete exact positive sense copy of the genome.
Nucleocapsid assembly occurs in the cytoplasm and is tightly linked to RNA synthesis. Nucleocapsids are enveloped by budding at the cell surface plasma membrane at sites containing virus envelope proteins. Members of the subfamily Paramyxovirinae contain 6–7 transcriptional elements that encode 7–11 proteins. Each element encodes a single mRNA with the sole exception of the P/V element. This element is transcribed into an exact-copy mRNA (P or V mRNA, depending on the genus) as well as into an alternative version in which the RNA transcriptase stutters on the template at an editing motif midway down the element. This results in the insertion of one or more pseudo-templated nucleotides (“RNA editing”) and shifts the reading frame to access an alternative ORF. The exact-copy and edited mRNAs synthesize two alternative proteins, P and V, which have identical amino-terminal domains but have different carboxy-terminal domains due to the frameshift. Other truncated, or chimeric, proteins (called I, W, or D, depending on the virus) can be produced by shifting into the third reading frame. The C ORF present in respiroviruses, henipa-viruses, and morbilliviruses overlaps the P ORF and can initiate synthesis at a non-AUG codon that is accessed by ribosomal choice or at alternative start codons in the same ORF.
Members of the subfamily Pneumovirinae have eight (Metapneumovirus) or 10 (Pneumovirus) transcriptional elements, each of which encodes one mRNA. Each mRNA has a single ORF, except for the M2 mRNA, which encodes two proteins from separate ORFs. There is overlap between the M2 and L transcriptional elements in some pneumoviruses (Figure 2), but these elements nonetheless give rise to separate mRNAs.
Antigenic properties
The attachment (HN, or H, or G) and fusion (F) proteins are of primary importance in inducing virus-neutralizing antibodies and immunity against reinfection. Antibodies to N and, variably, to other viral proteins also are induced by infection. Various proteins of members of the subfamily Paramyxovirinae have been reported to be broken into specific peptides that, when complexed to major histocompatibility glycoproteins, serve as recognition molecules for cytotoxic or helper T cells.
Biological properties
Paramyxoviruses have been conclusively identified only in vertebrates and mostly in mammals and birds, although they have recently also been detected in reptiles and fish. Most viruses have a narrow host range in nature, but in cultured cells they display a broad host range. Infection of cultured cells generally is lytic, but temperate or persistent infections in vitro are common. Other features of infection include the formation of inclusion bodies and syncytia. Cell surface molecules reported to serve as receptors for the attachment of respiroviruses and rubulaviruses include sialoglycoproteins and glycolipids. The cell surface proteins CD46 and SLAM 150 are major receptors for measles virus. Henipaviruses use ephrin B2 and B3 proteins as entry receptors. HRSV infection in vitro involves glucosaminoglycans. Nucleocapsids associate with viral membrane proteins at the plasma membrane and are enveloped by budding. Transmission is horizontal, mainly through airborne routes; no vectors are known. Paramyxovirus infection typically begins in the respiratory tract and may remain at that site (e.g., HRSV and HPIV) or may spread to secondary sites (e.g., lymphoid and endothelial tissues for MeV, the parotid gland, CNS and endothelial tissues for MuV or lung and CNS for HeV and NiV). In general, paramyxovirus infections are limited by, and eliminated by, host immunity. However, virus sometimes can be shed for periods of weeks or months in normal and, especially, immunocompromised individuals. Latent infection is unknown, and long-term persistent infection is known only for subacute sclerosing panencephalitis, a rare complication that involves defective measles virus and old dog distemper, which can involve persistence of defective or fully infectious virus for weeks or months in normal and, especially, immunocompromised individuals. The recurrence of neurological manifestations has also been noted in NiV patients more than 4 years after recovery from acute encephalitis.
Subfamily Paramyxovirinae
Distinguishing features
Members of the subfamily Paramyxovirinae have 6–7 transcriptional elements. Amino acid sequence relatedness is much greater within the subfamily than between subfamilies. Within the subfamily Paramyxovirinae, sequence relatedness between corresponding proteins is greater for N, M and L, with F and HN being somewhat less conserved and C and P being poorly conserved although the unique region of V that is not shared with P is highly conserved. The division of this subfamily into the five genera is consistent with phylogenetic grouping based on amino acid sequence relationships. Their nucleocapsids have diameters of 18 nm and a pitch of 5.5 nm; the length of the surface F and H/HN spikes is 8 nm. The genome length must be a multiple of 6 nt for efficient genome replication (the “rule of six”), perhaps reflecting the precise packing of nt by a nucleocapsid protein subunit. RNA editing of the P/V transcriptional element occurs for all members except human parainfluenza virus 1 (HPIV-1). One genus (Morbillivirus) lacks a neuraminidase activity and some viruses (canine distemper virus, phocine distemper virus and rinderperst virus) lack a detectable hemagglutinating activity. Viruses from the genus Henipavirus lack both neuraminidase and hemagglutinating activities.
Genus Rubulavirus
Type species Mumps virus
Distinguishing features
All species of the genus Rubulavirus have hemagglutination and neuraminidase activities. They share greater sequence relatedness within the genus than with members of other genera. For example, the N protein of HPIV-2 is 39–74% identical with that of MuV, PIV5, HPIV-4 and SV-41, compared to 18% and 24% identical with HPIV-1 and MeV. Some members (PIV5 and MuV) contain an extra transcriptional element (SH) between the F and HN loci (Figure 2). The unedited and edited versions of the mRNA from the P locus encode the V and P proteins, respectively. The intergenic sequences are of variable length. All members lack a C protein ORF. The rubulavirus P protein is substantially smaller than that of the respiroviruses or morbilliviruses. MuV and HIPV-2 are significant human pathogens.
Species demarcation criteria in the genus
HPIV-2 and human parainfluenza virus 4 (HPIV-4) represent distinct serotypes that lack significant cross-neutralization and cross-protection. HPIV-2, PIV5, and SV-41 exhibit considerable sequence relatedness and some antigenic relatedness, but these viruses can be distinguished on either basis (for example, the N protein of HPIV-2 is 57% or 74% identical to those of PIV5 or SV-41, respectively) as well as by host range: HPIV-2, HPIV-4a and HPIV-4b infect humans, PIV5 dogs, monkeys and humans, and SV-41 monkeys. They also have differences in their gene maps: PIV5 and MuV have an additional gene, SH and SV-41 lacks a functional transcription termination signal for the M gene and thus does not express a monocistronic M mRNA. HPIV-4 contains two antigenic subgroups (a and b) that are distinguished by differences in reactivity with monoclonal antibodies but are highly related by sequence – 84% and 95% identity for the HN and F protein, respectively – and should not be considered distinct species. MuV also does not exhibit significant cross-neutralization and cross-protection with other paramyxoviruses, and it is distinguished by its gene map (it contains an SH gene, found within this group only in PIV5), by sequence divergence (the MuV N protein shares 44% or less aa sequence identity with other rubulaviruses), and by its disease.
List of species in the genus Rubulavirus
| Human parainfluenza virus 2 |
|
|
| Human parainfluenza virus 2 V94 | [AF533010] | (HPIV-2-V94) |
| Human parainfluenza virus 4 |
|
|
| Human parainfluenza virus 4a M-25 | [AB543336] | (HPIV-4a-M25) |
| Human parainfluenza virus 4b 68-333 | [AB543337] | (HPIV-4b-68-33) |
| Mapuera virus |
|
|
| Mapuera virus BeAnn 370284 | [EF095490] | (MprPV-BeAnn370284) |
| Mumps virus |
|
|
| Mumps virus Miyahara | [AB040874] | (MuV-Miy) |
| Parainfluenza virus 5 |
|
|
| Parainfluenza virus 5 W3A | [AF052755] | (PIV5-W3A) |
| (Simian virus 5) |
| (SV5) |
| Porcine rubulavirus |
|
|
| Porcine rubulavirus LPMV | [BK005918] | (PorPV-LPMV) |
| (La-Piedad-Michoacan-Mexico virus) |
| (LPMV) |
| Simian virus 41 |
|
|
| Simian virus 41Toshiba/Chanock | [X64275] | (SV-41-Tos/Ch) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Rubulavirus but have not been approved as species
| Tioman virus | [AF298895] | (TioPV) |
| Menangle virus | [AF326114] | (MenPV) |
Genus Avulavirus
Type species Newcastle disease virus
Distinguishing features
All species of the genus Avulavirus have hemagglutinin and neuraminidase activities. They share greater sequence relatedness within the genus than with members of other genera but are closely related to the rubulaviruses. The major distinguishing feature between avulaviruses and rubulaviruses is that for avulaviruses the exact copy of the P/V mRNA encodes P and the edited form encodes V, and none of the avulaviruses have an SH gene. The intergenic sequences are of variable length. All members lack a C protein ORF. The avulavirus P protein is substantially smaller than that of the respiroviruses or morbilliviruses. These viruses, especially NDV, include significant avian pathogens.
Species demarcation criteria in the genus
There are many strains of NDV (virulent and avirulent) for chickens that have been extensively analyzed. The many other avian paramyxoviruses are antigenically related and are not well studied. These are serotypes defined by hemagglutination-inhibition tests, although weak interactions occur between the types. Also, each serotype has a distinct pattern of electrophoretically-separated polypeptides.
List of species in the genus Avulavirus
| Avian paramyxovirus 2 |
|
|
| Avian paramyxovirus 2 (Yucaipa) | [EU338414] | (APMV-2-Yucaipa) |
| Avian paramyxovirus 3 |
|
|
| Avian paramyxovirus 3 449/75 | [EU403085] | (APMV-3-449/75) |
| Avian paramyxovirus 4 |
|
|
| Avian paramyxovirus 4KR/YJ/06 | [EU877976] | (APMV-4-YJ/06) |
| Avian paramyxovirus 5 |
|
|
| Avian paramyxovirus 5 (Kunitachi) | [GU206351] | (APMV-5-Kunitachi) |
| Avian paramyxovirus 6 |
|
|
| Avian paramyxovirus 6 4440/2003 | [EF569970] | (APMV-6-4440/2003) |
| Avian paramyxovirus 7 |
|
|
| Avian paramyxovirus 7 Tennessee 4/75 | [FJ231524] | (APMV-7-Tennessee) |
| Avian paramyxovirus 8 |
|
|
| Avian paramyxovirus 8 Delaware 1053/76 | [FJ215863] | (APMV-8-Delaware) |
| Avian paramyxovirus 9 |
|
|
| Avian paramyxovirus 9 New York/22/1978 | [EU910942] | (APMV-9-NY/22/78) |
| Newcastle disease virus |
|
|
| Newcastle disease virus LaSota | [AF077761] | (NDV-La Sota) |
| (Avian parainfluenza virus 1) |
| (APMV-1) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Avulavirus but have not been approved as species
None reported.
Genus Respirovirus
Type species Sendai virus
Distinguishing features
Member viruses of the genus Respirovirus possess a hemagglutinin and a neuraminidase. These viruses have six transcriptional elements. Unedited P mRNA encodes the P protein, whereas insertion of a G nucleotide in P mRNA transcripts accesses the V ORF. All members encode a C protein from a separate ORF in the P/V mRNAs. Amino acid sequence relatedness within the genus ranges from low to high, depending on the protein, and always is higher than in comparisons with other genera. For example, within respiroviruses the N protein is 88% identical comparing HPIV-1 to SeV, its “murine counterpart”, and 63% identical compared to HPIV-3. Between genera, the N protein is 18% identical between HPIV-1 (Respirovirus) and the N protein of MuV or HPIV-2 or HPIV-4 (Rubulavirus), and 21% compared with that of MeV (Morbillivirus). HPIV-1 and HPIV-3 are significant agents of respiratory tract disease.
Species demarcation criteria in the genus
HPIV-1 and HPIV-3 represent distinct serotypes defined by a lack of significant cross-neutralization and cross protection. Their gene maps are similar but not identical (editing by HPIV-3 results in a unique D protein not seen elsewhere in Paramyxoviridae, but does not access the V coding sequence, whereas HPIV-1 is the only member of the subfamily that lacks editing). They share low to high sequence relatedness among the various proteins (see above). SeV, a close relative of HPIV-1, is found predominantly as a pathogen of laboratory mice. However, this virus was isolated from the lungs of infants during a fatal epidemic of newborn pneumonitis in Japan in 1952 and this virus is not found in wild mice in either Japan or the USA. SeV is distinguished from HPIV-1 by host range: specifically, HPIV-1 is permissive and pathogenic in humans whereas in mice it grows poorly or not at all and is nonpathogenic. Conversely, SeV is highly permissive, transmissible and pathogenic for mice. The two viruses have considerable sequence relatedness (see above) and antigenic similarity, but also can be clearly distinguished on either basis; also, HPIV-1 lacks editing and a V protein. Bovine parainfluenza virus 3 (BPIV-3) is a close relative of HPIV-3, but differs by their host ranges, which overlap but exhibit specificity. For example, in humans HPIV-3 replicates efficiently, is easily transmissible and causes disease, whereas BPIV-3 is highly attenuated, nonpathogenic and poorly transmissible. Furthermore, compared to HPIV-3, BPIV-3 is restricted 100- to 1000-fold in Old World primates. HPIV-3 and BPIV-3 exhibit considerable genetic and antigenic similarity, but also can clearly be distinguished on either basis. For example, HPIV-3 and BPIV-3 are 25% related antigenically by reciprocal cross-neutralization and hemagglutination inhibition studies. Also, BPIV-3 makes a V protein whereas it is not clear whether HPIV-3 can. Simian virus 10 (SV10; also known as simian agent 10) is a hemagglutinating virus that was recovered from the mouth of a samango monkey (Cercopithecus mitis). Sequence analysis indicates that SV10 and HPIV3 are essentially indistinguishable, implying that the species Simian virus 10 should be abolished and SV10 viewed as being a monkey variant of the species Human parainfluenza virus 1. With the exception of Simian virus 10, each species represents a significant pathogen in its respective host.
List of species in the genus Respirovirus
| Bovine parainfluenza virus 3 |
|
|
| Bovine parainfluenza virus 3 Kansas 15626/84 | [AF178654] | (BPIV-3-Kansas) |
| Human parainfluenza virus 1 |
|
|
| Human parainfluenza virus 1 Washington/1964 | [AF457102] | (HPIV-1-Washington) |
| Human parainfluenza virus 3 |
|
|
| Human parainfluenza virus 3 14702 | [EU424062] | (HPIV-3-14702) |
| Sendai virus |
|
|
| Sendai virus Nagoya | [AB195968] | (SeV-Nagoya) |
| (Murine parainfluenza virus 1) |
|
|
| Simian virus 10 |
|
|
| Simian virus 10 | [HM583801] | (SA-10 or SV-10) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Respirovirus but have not been approved as species
None reported.
Genus Henipavirus
Type species Hendra virus
Distinguishing features
The two species of the genus Henipavirus have an attachment protein (G) that lacks hemagglutinating and neuraminidase activities. They share greater sequence relatedness within the genus than with members of other genera. The major distinguishing features between henipaviruses and other paramyxoviruses are (i) the long 5′- and 3′-UTRs in the mRNAs and (ii) a genome that as a result is approximately 3000 nt or more longer than other members of the family Paramyxoviridae. The unedited P mRNA encodes P protein. The edited P mRNA encodes V protein. The intergenic sequences are three nt at each gene junction. Both members encode a C protein ORF. Both HeV and NiV are indigenous to fruit bats. Each species has been associated with limited outbreaks with high mortality in domesticated animals and humans. Different from most paramyxoviruses, henipaviruses have a wide host range from bats to pigs, horses and humans. The identification of the highly conserved ephrin B2 as the main functional receptor for both HeV and NiV and the widespread occurrence of the molecule in vertebrates, particularly in arterial, but not venous, endothelial cells, in the smooth muscle of the tunica media and in neurons, provide an explanation for the wide host range of henipaviruses and the systemic nature of the infections they cause.
Species demarcation criteria in the genus
HeV and NiV are antigenically distinct and are distinct by genome sequence and geographic location. HeV has been detected exclusively in Australian flying foxes whereas NiV or anti-NiV antibodies have been detected in fruit bats from Indonesia, Malaysia, Thailand, Cambodia, India and Bangladesh, to Madagascar and West Africa. The two viruses cross-neutralize and have considerable sequence relatedness, but also can be distinguished on either basis
List of species in the genus Henipavirus
| Hendra virus |
|
|
| Hendra virus Hendra | [AF017149] | (HeV-Hendra) |
| Nipah virus |
|
|
| Nipah virus Malaysia | [AF212302] | (NiV-Malaysia) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Henipavirus but have not been approved as species
None reported.
Genus Morbillivirus
Type species Measles virus
Distinguishing features
Members of the genus Morbillivirus lack a neuraminidase activity. Member viruses exhibit greater amino acid sequence relatedness within the genus than with other genera. They have an identical gene order, number of transcriptional elements and size of intergenic sequences with members of the genus Respirovirus (Figure 2). All morbilliviruses have a P/C/V transcription unit with RNA editing like respiroviruses and henipaviruses, namely the templated exact-copy mRNA encodes P and the predominant edited mRNA form (1 G added) encodes V. All morbilliviruses produce both intracytoplasmic and intranuclear inclusion bodies containing nucleocapsid-like structures. Viruses cross-react in serological tests. Sialic acid does not appear to be a receptor for morbilliviruses. Narrow host-range distribution of receptor defines susceptibility of organisms to infection. For MeV one receptor is CD46 and another CD150. CD150 also appears to be a receptor for CDV and RPV, which have a preference for canine and bovine CD150 respectively. Each species is a significant cause of disease in its respective host.
Species demarcation criteria in the genus
The morbilliviruses are distinguished by host range, genetic (sequence) and antigenic differences. There is a low to moderate degree of sequence relatedness between members, depending on the protein (for example, the N protein of MeV is 65% related to that of CDV, viruses that represent two branches of the genus). Cross-neutralization and cross-protection also occurs between members of the genus, although members can also be distinguished on that basis. MeV infects primates, CDV infects members of the order Carnivora, and RPV and peste-des-petits-ruminants virus (PPRV) infect members of the order Artiodactyla (even-toed ungulates), especially ruminants and swine. PPRV is distinguished from RPV by sequence analysis (the N protein of PPRV shares 68–72% identity with that of MeV, RPV or CDV), and because it does not readily infect cattle. Phocine distemper virus (PDV) is most closely related to CDV and is distinguished by host range and sequence divergence. Two cetacean morbilliviruses have been described, dolphin morbillivirus (DMV) and porpoise morbillivirus (PMV), but these are closely related and now are considered to be members of a single species, now named Cetacean morbillivirus. Members of this species are most closely related to RPV and MeV: these viruses are distinguished by host range and sequence divergence.
List of species in the genus Morbillivirus
| Canine distemper virus |
|
|
| Canine distemper virus 007Lm | [AB474397] | (CDV-007Lm) |
| Cetacean morbillivirus virus |
|
|
| Cetacean morbillivirus virus Dolphin | [AJ608288] | (CeMV-dolphin) |
| Measles virus |
|
|
| Measles virus Edmonston | [AF266288] | (MeV-Edmonston) |
| Peste-des-petits-ruminants virus |
|
|
| Peste-des-petits-ruminants virus ICV89 | [EU267273] | (PPRV-ICV89) |
| Phocine distemper virus |
|
|
| Phocine distemper virus Ulster 88 | [D10371*, Y09630*] | (PDV-Ulster88) |
| Seal distemper virus |
|
|
| Rinderpest virus |
|
|
| Rinderpest virus RBOK | [Z30697] | (RPV-RBOK) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.*Partial sequences contributing to the complete genome.
List of other related viruses which may be members of the genus Morbillivirus but have not been approved as species
None reported.
Subfamily Pneumovirinae
Distinguishing features
Members differ from those of the subfamily Paramyxovirinae in several features: (a) possession of 8–10 separate transcriptional elements; (b) smaller average ORF size; (c) possession of an additional nucleocapsid-associated protein (M2-1, formerly called 22K) and an RNA regulatory protein (M2-2); (d) extensive O-linked glycosylation of the G protein; (e) the P mRNA has a single ORF and does not have RNA editing; (f) a lack of amino acid sequence relatedness with members of the subfamily Paramyxovirinae except for a low level in the F and L proteins; and (g) differences in nucleocapsid diameter (13–14 nm compared with 18 nm in the subfamily Paramyxovirinae), nucleocapsid pitch (7 nm) and length of glycoprotein spikes (10–14 nm); and (h) lack of a “rule of six” governing the nucleotide length. Species also lack a neuraminidase and a hemagglutinin except in the case of murine pneumonia virus (MPV) (formerly pneumonia virus of mice [PVM]), which has a hemagglutinin. The G attachment protein is structurally unrelated to the HN, H or G proteins of the subfamily Paramyxovirinae and exhibits a high level of inter-strain diversity: only 53% identity among HRSV isolates, 21–30% identity between human and non-human respiratory syncytial viruses, and 37% identity between HMPV strains.
Genus Pneumovirus
Type species Human respiratory syncytial virus
Distinguishing features
Pneumoviruses are distinguished from metapneumoviruses by (i) the presence of the NS1 and NS2 genes, (ii) the SH, G, F and M2 genes being in the order SH-G-F-M2 as opposed to F-M2-SH-G for metapneumoviruses, (iii) a greater genome length (15,190–15,225 nt compared to 13,280–13,378 nt), and (iv) a higher degree of nucleotide and amino acid sequence relatedness within the genus than between genera.
Species demarcation criteria in the genus Pneumovirus
HRSV and MPV are distinguished by host range (humans versus mice) and a lack of cross-neutralization. Amino acid sequence relatedness between these two viruses varies from undetectable to intermediate, depending on the protein (for example, the NS1 and NS2 proteins lack demonstrable relatedness, whereas the N or F proteins share 60% or 40% identity, respectively). Their gene maps differ only in the absence of the M2/L gene overlap in MPV. Bovine respiratory syncytial virus (BRSV) differs from HRSV in host range, specifically cattle versus humans, but the difference is not absolute. For example, both viruses grow efficiently in cultured human or bovine cells, although some specificity may be evident. In chimpanzees, HRSV replicates efficiently, is transmissible and is pathogenic, whereas BRSV is very attenuated and non-pathogenic. The two viruses share considerable sequence and antigenic relatedness, but also can clearly be distinguished on either basis. For example, the N or F proteins are each 81% identical between BRSV and HRSV, compared to 96% or 89% identical, respectively, among different HRSV strains. Antiserum against one virus will cross-neutralize the other with a 6- to 64-fold reduction in efficiency. There are two antigenic subgroups of HRSV, called A and B, which exhibit aa sequence identity ranging from 96% (N) to 53% (G), and which are approximately 50% or 5% related antigenically in the F or G protein, respectively, with the overall difference in reciprocal cross-neutralization being up to four-fold. Comparable antigenic dimorphism also may exist for BRSV.
List of species in the genus Pneumovirus
| Bovine respiratory syncytial virus |
|
|
| Bovine respiratory syncytial virus ATCC51908 | [AF295543] | (BRSV-ATCC51908) |
| Human respiratory syncytial virus |
|
|
| Human respiratory syncytial virus A2 | [M74568] | (HRSV-A2) |
| Human respiratory syncytial virus B1 | [AF013254] | (HRSV-B1) |
| Human respiratory syncytial virus S2 | [U39662] | (HRSV-S2) |
| Murine pneumonia virus |
|
|
| Murine pneumonia virus 15 (formerly Pneumonia virus of mice [PVM]) | [AY729016] | (MPV-15) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Pneumovirus but have not been approved as species
Caprine and ovine strains of BRSV also have been described but might represent, with BRSV, a subgroup of ruminant strains rather than different species.
Genus Metapneumovirus
Type species Avian metapneumovirus
Distinguishing features
The relative placements of SH-G versus F-M2 in the gene order are reversed as compared to pneumoviruses. NS1 and NS2 genes are absent in pneumoviruses, and the genome is nearly 2000 nt shorter. The intergenic regions are longer (up to 190 nt compared with 57 nt). The extent of sequence relatedness is greater within than between genera.
Species demarcation criteria in the genus
Metapneumovirus species are distinguished on the basis of having an avian or human host. Interestingly, the sequence diversity between avian and human isolates is less than that between certain avian isolates, and thus sequence relatedness is not a reliable distinguishing feature.
List of species in the genus Metapneumovirus
| Avian metapneumovirus |
|
|
| Avian metapneumovirus Colorado (formerly Turkey rhinotracheitis virus and Avian pneumovirus) | [AY590688] | (AMPV-Colorado) |
| Human metapneumovirus |
|
|
| Human metapneumovirus 00-1 | [AF371337] | (HMPV-00-1) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Metapneumovirus but have not been approved as species
None reported.
List of other related viruses which may be members of the family Paramyxoviridae but have not been approved as species
| Fer-de-Lance virus | [AY141760] | (FdlPV) |
| Atlantic salmon paramyxovirus | [EF646380] | (AsaPV) |
| Pacific salmon paramyxovirus | [AY536862] | (PsaPV) |
| Nariva virus | [EF095490] | (NarPV) |
| Mossman virus | [AY286409] | (MosPV) |
| Salem virus | [AF237881] | (SalPV) |
| Tupaia paramyxovius | [AF079780] | (TupPV) |
| J-virus | [AY900001] | (JPV) |
| Beilong virus | [DQ100461] | (BeiPV) |
Phylogenetic relationships within the family
The literature on the relationships of members of the subfamily Paramyxovirinae is consistent with the phylogeny (see Figure 3).
Similarity with other taxa
The member viruses of the family Paramyxoviridae have a similar strategy of gene expression and replication and gene order to those of other families in the order Mononegavirales, specifically the families Rhabdoviridae and Filoviridae.
Derivation of names
Avula: from avian rubula virus.
Henipa: from Hendra and Nipah viruses.
Meta: from Greek meta, “after”.
Morbilli: from Latin morbillus, diminutive of morbus, “disease”.
Ortho: from Greek orthos, “straight”.
Paramyxo: from Greek para, “by the side of”, and myxa, “mucus”.
Pneumo: from Greek pneuma, “breath”.
Respiro: from Latin respirare, “respire, breathe”.
Rubula: Rubula inflans – old name for mumps.
Further reading
Carbone and Rubin, 2007 K.M. Carbone, S.A. Rubin, D.M. Knipe, P.M. Howley, Mumps virusFields Virology. In: D.M. Knipe, P.M. Howley, Fields Virology. Lippincott Williams and Wilkins, Philadelphia, PA20071527–1550.
Collins and Crowe, 2007 P.L. Collins, J.E. Crowe, D.M. Knipe, P.M. Howley, Respiratory syncytial virus and metapneumovirusFields Virology. In: D.M. Knipe, P.M. Howley, Fields Virology. Lippincott Williams and Wilkins, Philadelphia, PA200711601–11646.
Eaton et al., 2007 B.T. Eaton, J.S. Mackenzie, L.-F. Wang, D.M. Knipe, P.M. Howley, HenipavirusesFields Virology. In: D.M. Knipe, P.M. Howley, Fields Virology. Lippincott Williams and Wilkins, Philadelphia, PA20071587–11600.
Falk et al., 2008 K. Falk, W.N. Batts, A. Kvellestad, G. Kurath, J. Wiik-Nielsen, J.R. Winton, Molecular characterisation of Atlantic salmon paramyxovirus (ASPV): A novel paramyxovirus associated with proliferative gill inflammation. Virus Res. 133 (2008) 218–227.
Griffin, 2007 D.E. Griffin, D.M. Knipe, P.M. Howley, Measles virusFields Virology. In: D.M. Knipe, P.M. Howley, Fields Virology. Lippincott Williams and Wilkins, Philadelphia, PA20071551–1586.
Kalter et al., 1980 S.S. Kalter, D. Ablashi, C. Espana, R.L. Heberling, R.N. Hull, E.H. Lennette, H.H. Malherbe, S. McConnell, D.S. Yohn, Simian virus nomenclature, 1980. Intervirology. 13 (1980) 317–330.
Karron and Collins, 2007 R.A. Karron, P.L. Collins, D.M. Knipe, P.M. Howley, Parainfluenza virusesFields Virology. In: D.M. Knipe, P.M. Howley, Fields Virology. Lippincott Williams and Wilkins, Philadelphia, PA200711497–11526.
Lamb and Parks, 2007 R.A. Lamb, G.D. Parks, D.M. Knipe, P.M. Howley, Paramyxoviridae: the viruses and their replicationFields Virology. In: D.M. Knipe, P.M. Howley, Fields Virology. Lippincott Williams and Wilkins, Philadelphia, PA20071449–1496.
Li et al., 2006 Z. Li, M. Yu, H. Zhang, D.E. Magoffin, P.J.M. Jack, A. Hyatt, H.-Y. Wang, L.-F. Wang, Beilong virus, a novel paramyxovirus with the largest genome of non-segmented negative-stranded RNA viruses. Virology. 346 (2006) 219–228.
Wang et al., 2003 L.-F. Wang, K.B. Chua, M. Yu, B.T. Eaton, Genome diversity of emerging paramyxoviruses. Curr. Genom. 4 (2003) 263–273.
Wang and Eaton, 2001 L.-F. Wang, B.T. Eaton, Emerging paramyxoviruses. Infect. Dis. Rev. 3 (2001) 52–69.
Contributed by
Wang, L.-F., Collins, P.L., Fouchier, R.A.M., Kurath, G., Lamb, R.A., Randall, R.E. and Rima, B.K.
Figures
Figure 1 (Right) Negative contrast electron micrographs of intact parainfluenza virus 5 (PIV5, previously known as simian virus 5 [SV5]) particles (genus Rubulavirus) (top) and the PIV5 nucleocapsid after detergent lysis of virions (bottom) (courtesy of G.P. Leser and R.A. Lamb). The bars represent 100 nm. (Left top and bottom) Schematic diagrams of PIV5 particles in cross-section (N) (formerly NP), nucleocapsid; P, phospho-protein; L, large polymerase protein; V, cysteine rich protein that shares its N-terminus with P sequence and for PIV5 is found in virions; M, matrix or membrane protein; F, fusion protein; HN, hemagglutinin-neuraminidase; SH, small hydrophobic protein). (Adapted from Kingsbury, D.W. (1990). Paramyxoviridae: the viruses and their replication. In: Virology, 2nd edn (B.N. Fields and D.M. Knipe, Eds.), Raven Press, New York; and from Scheid, H. (1987). In: Animal Virus Structure (M.V. Nermut, and A.C. Steven, Eds.), Elsevier, Amsterdam; with permission.)
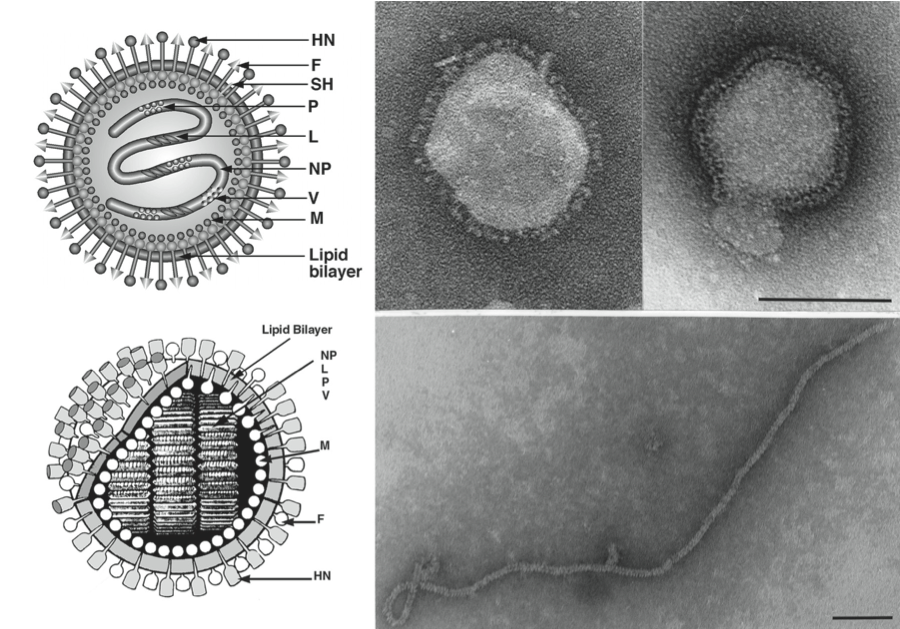
Figure 2 Maps of genomic RNAs (3-to-5) of viruses in the family Paramyxoviridae. Viruses were selected from the seven established genera as well as a group of unassigned viruses, which significantly increased the genetic diversity of the family. Each box represents a separately encoded mRNA; multiple distinct ORFs within a single mRNA are indicated by slashes. The M2 mRNA of members of subfamily Pneumovirinae has two overlapping ORFs, M2-1 and M2-2 (not shown). The lengths of the boxes are approximately to scale although the intervening or preceding sequences are not to scale. The D ORF present in the respirovirus HPIV3 is not shown. Certain viruses give rise to additional proteins by the utilization of secondary translational start sites within some of the ORFs: these are not shown. In HPIV1 and HPIV3 of the genus Respirovirus the V ORF may be a non-expressed relic. In the genus Rubulavirus some species lack the SH gene. In the genus Pneumovirus, HRSV has a transcriptional overlap at M2 and L (staggered boxes). There are conserved trinucleotides that serve as intergenic sequences for the respiroviruses, henipaviruses and morbilliviruses. For rubulaviruses, avulaviruses, pneumoviruses and meta-pneumoviruses, the intergenic sequences are variable (1190 nt long). In the group of unassigned new viruses, all of them have a 3-nt intergenic region similar to those observed in the genera Morbillivirus, Respirovirus and Henipavirus. However, the genome sizes of these new viruses vary significantly from 15,378 nt to 19,212 nt. There are also novel genes present in these viruses (such as the U gene in FedPV and the TM gene in BeiPV) which have never been seen in previously known paramyxoviruses.

Figure 3 Phylogenetic analysis of the L proteins of members of the family Paramyxoviridae. Phylogenetic analysis using MEGA4.1 was performed on the aa sequence of L proteins from various members of the family Paramyxoviridae. The tree shown was based on maximum parsimony; however, analysis of the same data using maximun likelihood produced a tree with nearly identical topology (data not shown).

