Family: Filoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
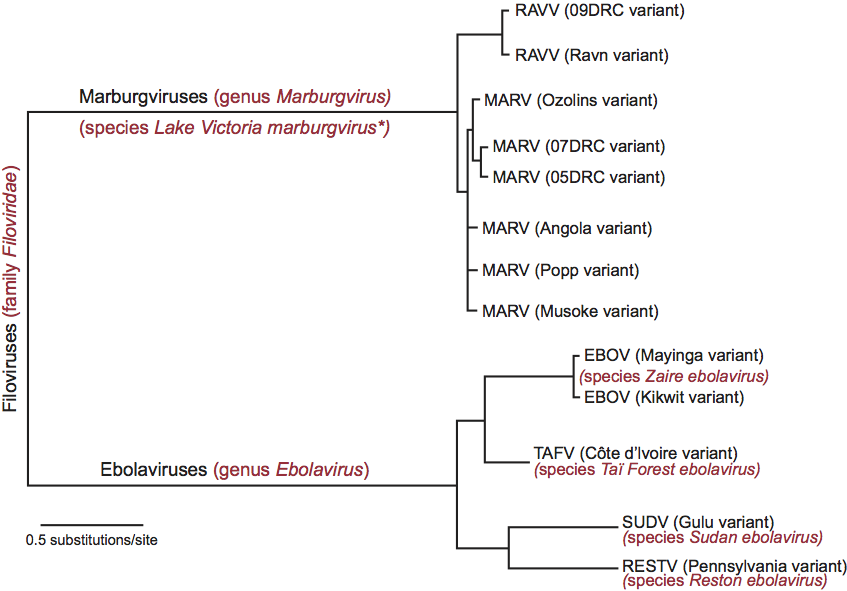
Virions are bacilliform in shape, but particles can also appear as branched, circular, U or 6-shaped and long filamentous forms. Spherical forms are rare to absent (Figure 1). This morphology is unusual for mammalian viruses and, thus, is considered characteristic for members of the family. Virions vary greatly in length but show a uniform width of about 80 nm. Family members differ in length of virions, but seem to be very similar in morphology. Peak infectivity has been associated with particles of about 665 nm in the case of marburgviruses and about 805 nm in the case of ebolaviruses.
Virions are composed of a central core formed by a nucleocapsid or ribonucleoprotein (RNP) complex, surrounded by a lipid envelope derived from the host-cell plasma membrane. Electron micrographs reveal an axial channel (about 10–15 nm in width) surrounded by a central dark layer (about 20 nm in width) and an outer helical layer (about 50 nm in width) with cross-striations of about 5 nm intervals (Figure 1). Spikes about 7 nm in diameter and spaced at intervals of about 10 nm are seen as globular structures on the surface of virions.
Physicochemical and physical properties
Virions have a molecular weight of about 3.82×108. The buoyant density of virions in potassium tartrate is about 1.14 g cm−3. The S20,w of bacilliform particles is 1.40S, but higher for longer particles. The nucleocapsid has a buoyant density in CsCl of about 1.32 g cm−3.
Nucleic acid
Filovirus genomes are nonsegmented, single stranded, linear RNA molecules of negative polarity with lengths of about 19 kb: marburgvirus genomes are 19.1 kb in length and ebolavirus genomes are 18.9 kb. The Mr of a genomic RNA is about 4.2×106 and the genome represents about 1.1% of the total virion mass. Genomic RNA is not polyadenylated at the 3′ end and there is no evidence for a 5′-terminal cap structure or a covalently-linked protein. Full-length nucleotide sequences of the genomes of members of all filovirus species have been determined.
Proteins
Filovirus RNP complexes are composed of a genomic RNA molecule and four virion-associated structural proteins: NP (nucleoprotein), VP35 (RNA-dependent RNA polymerase cofactor), VP30 (transcriptional activator), and L (RNA-dependent RNA polymerase). The three remaining structural proteins are membrane-associated: GP1,2 (spike glycoprotein) is a type 1 transmembrane and class I fusion protein, whereas the two non-glycosylated proteins VP40 and VP24 (primary and secondary matrix proteins) are associated with the inner side of the virion membrane. The sizes and functions of the proteins are shown in Table 1.
Table 1 Filovirus structural proteins
| Protein | Encoding gene | Function | Marburg virus (kDa) A/B | Ebola virus (kDa) A/B |
| NP | 1 (NP) | Nucleoprotein | 77.9/96.0 | 83.3/104.0 |
| VP35 | 2 (VP35) | RNA-dependent RNA polymerase cofactor (paramyxovirus P protein analog) | 31.0/32.0 | 38.8/35.0 |
| VP40 | 3 (VP40) | Primary matrix protein (paramyxovirus M protein analog) | 31.7/38.0 | 35.3/40.0 |
| GP1,2 | 4 (GP) | Spike glycoprotein (paramyxovirus G protein analog) | 74.8/170.0 | 74.5/140.0 |
| VP30 | 5 (VP30) | Transcriptional activator | 31.5/28.0 | 29.7/30.0 |
| VP24 | 6 (VP24) | Secondary matrix protein | 28.8/24.0 | 28.3/24.0 |
| L | 7 (L) | RNA-dependent RNA polymerase | 267.2/>200.0 | 252.7/>200.0 |
A, size calculated from the deduced amino acid sequences of the corresponding ORFs; B, size estimated from SDS-PAGE analysis; a complete set of data is not yet available for ebolaviruses other than EBOV.
Lipids
The viral envelope is derived from host cell membranes and is considered to have a lipid composition similar to that of the host-cell plasma membrane.
Carbohydrates
The spike glycoproteins of filoviruses are highly glycosylated with N-linked glycans of the complex, hybrid and oligomannosidic type, and O-linked glycans of the neutral mucin type. Glycans constitute >50% of the GP1,2 total mass. Ebola virus (EBOV) GP1,2 is sialylated to higher levels than Marburg virus (MARV) GP1,2, and in certain cell lines the GP1,2 of MARV completely lacks sialic acid. The GP1,2 proteins of marburgviruses and ebolaviruses are acylated at their C-termini and form homotrimers.
Genome organization and replication
Filovirus genomes are characterized by the gene order: 3′-NP-VP35-VP40-GP-VP30-VP24-L-5′ (Figure 2). The extragenic sequences at the extreme 3′ (leader) and 5′ (trailer) ends of the genomes are conserved. They demonstrate a significantly high complementarity at their very ends. Genes are flanked by conserved transcriptional initiation and termination (polyadenylation) sites. Those sites contain the highly conserved pentamer 3′-UAAUU-5′ Most genes are separated by non-conserved intergenic sequences, but some genes overlap. Most of these overlaps are extremely short and limited to the highly conserved pentamer. There is a single gene overlap in marburgvirus genomes, but several (2–3) gene overlaps in ebolavirus genomes. The functional significance of these short overlaps is unclear, but transcriptional attenuation of the downstream gene has been postulated for other viruses of the order Mononegavirales. In addition, most genes possess relatively long 3′- and 5′-noncoding regions. In contrast to MARV, the GP genes of ebolaviruses possess three overlapping ORFs that can be joined through transcriptional polymerase stuttering (Figure 2).
The replication strategy of filoviruses is not well studied. Ultrastructural studies indicate an association of viral particles with coated pits for the initiation of infection, suggesting that filoviruses enter cells by endocytosis; molecular studies in part confirm this but are contradictory in terms of which endocytosis pathway is used. The spike glycoprotein mediates receptor binding and subsequent fusion. C-type lectins and integrins have been described as potential virion attachment factors, but the cell surface receptor remains elusive. Uncoating is presumed to occur in a manner analogous to that of other mononegaviruses. Transcription and genome replication take place in the cytoplasm and, in general, follow the models for members of the families Paramyxoviridae and Rhabdoviridae. Transcription starts at the conserved start site and polyadenylation occurs at a stretch of uridine residues within the termination site. The 5′-terminal non-coding sequences favor hairpin-like structures for all sgRNAs (mRNAs). Replication involves the synthesis of full-length positive-strand copies (antigenomes). During infection, massive amounts of nucleocapsids accumulate intracellularly and form intracytoplasmic inclusion bodies. Virions are released via budding from plasma membranes. The expression strategy of ebolaviral GP genes is distinct from that of marburgviral GP genes and involves transcriptional editing (Figure 2). The primary product of the unedited transcript (ORF1) yields a non-structural glycoprotein (pre-sGP), which is proteolytically cleaved to two secreted proteins, sGP and Δ-peptide. Editing results in the expression of either full-length GP1,2 or a secondary secreted glycoprotein (ssGP). sGP, GP1,2 and ssGP share the N-terminal 295 amino acid residues, but differ in their C-termini and, because of that, in quarternary structure.
Antigenic and genetic properties
Filovirus infectivity is poorly neutralized in vivo, but at least two potent neutralizing antibodies have been described that are potent in vitro (Sudan virus: 16F6; Ebola virus: KZ52). There is only limited antigenic cross-reactivity between marburgvirions and ebolavirions. Marburgvirus and ebolavirus GP genes differ by >57%, and phylogenetic analysis clearly separates marburgviruses and ebolaviruses, justifying the establishment of two filovirus genera (Marburgvirus and Ebolavirus) (Figure 3). One marburgvirus (Marburg virus, species Lake Victoria marburgvirus*) and four distinct ebolaviruses (Reston virus, species Reston ebolavirus; Sudan virus, species Sudan ebolavirus; Taï Forest virus, species Taï Forest ebolavirus; and Ebola virus, species Zaire ebolavirus) have been identified. They differ from one another by 37–41% at the nucleotide level. At least five genetic marburgvirus lineages exist. Virus genomes of four lineages (Marburg virus) differ from each other by 0–7.4%. Genomes from viruses of the fifth lineage (Ravn virus) reach 21% nucleotide difference compared to the others. The variation in genomic sequences has been shown to be low among isolates of individual filoviruses: <2%, for instance, in the case of Ebola virus isolates. There seems to be less or even no genetic variability between isolates from different patients of single outbreaks. All data indicate a remarkable degree of genetic stability over time.
Biological properties
Filoviruses are endemic in Central Africa in an area approximately between the 10th parallel north and south of the Equator as indicated by the locations of known outbreaks, and in the Philippines (Reston virus only). Filoviruses are the only mononegaviruses that cause viral hemorrhagic fevers in primates.
The natural reservoirs of filoviruses remain to be identified, but infectious marburgviruses have been isolated from Egyptian rousettes (Rousettus aegyptiacus) and infectious Reston virus has been isolated from domestic pigs (Sus scrofa). There is no known connection of virus spread with any vector. The usual transmission pattern seen with large outbreaks of filovirus disease in humans begins with a focus of infection that disseminates to a number of contacts. Secondary and subsequent episodes of disease occur following close contact with patients; such infections usually occur in family members or medical personnel. The major route of interhuman transmission of filoviruses requires direct contact with blood, body fluids, or injured skin (ritual embalming). Usage of contaminated syringes and needles is the main source for nosocomial infections. In the laboratory, nonhuman primates, mice, guinea pigs, hamsters and domestic pigs have been infected experimentally, but infection of rodents requires sequential adaptation.
Genus Marburgvirus
Type species Lake Victoria marburgvirus*
Distinguishing features
- Almost no antigenic cross-reactivity of marburgvirions with ebolavirions.
- Virion length is about 665 nm compared to about 805 nm for ebolavirions.
- Single gene overlap compared to several (2–3) overlaps in genomes of ebolaviruses.
- Spike glycoprotein expression does not involve editing during transcription of gene four.
- Gene four encodes only one protein, the spike glycoprotein GP1,2.
- Genomes differ from those of ebolaviruses by ≥50% at the nucleotide level.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Marburgvirus
| Lake Victoria marburgvirus* |
|
|
| Marburg virus - Musoke, Kenya, 1980 | [Z12132=NC_001608] | (MARV-Mus) |
| Ravn virus - Ravn, Kenya, 1987 | [DQ447649] | (RAVV-Rav) |
Species names are in italic script; names of strains and isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
Other related viruses which may be members of the genus Marburgvirus but have not been approved as species
None reported.
Genus Ebolavirus
Type species Zaire ebolavirus
Distinguishing features
- Almost no antigenic cross-reactivity of ebolavirions with marburgvirions.
- Virion length is about 805 nm compared to about 665 nm for marburgvirions.
- Several (2–3) gene overlaps compared to a single overlap in genomes of marburgviruses.
- Spike glycoprotein expression involves editing during transcription of gene four.
- Gene four encodes four proteins, the spike glycoprotein GP1,2 and three soluble glycoproteins (sGP/Δ-peptide and ssGP).
- Genomes differs from those of marburgviruses by ≥50% at the nucleotide level.
Species demarcation criteria in the genus
Members of different species in the genus may be distinguished on the basis of glycoprotein gene sequence differences (≥30% amino acid difference), whole genome differences (≥30% nucleo-tide difference), cross-protection data (where available), number of gene overlaps (two in Reston virus genomes, but three in all others) and differences in geographic origin (Taï Forest virus: Côte d’Ivoire; Reston virus: Philippines; Sudan virus: Sudan, Uganda; Ebola virus: Democratic Republic of the Congo, Gabon, Republic of the Congo).
List of species in the genus Ebolavirus
| Taï Forest ebolavirus |
|
|
| Taï Forest virus - Côte d’Ivoire, Côte d’Ivoire, 1994 | [FJ217162] | (TAFV-Côt) |
| Reston ebolavirus |
|
|
| Reston virus - Pennsylvania, USA, 1989 | [AF522874=NC_004161] | (RESTV-Pen) |
| Sudan ebolavirus |
|
|
| Sudan virus - Boniface, Sudan, 1976 | [FJ968794] | (SUDV-Bon) |
| Zaire ebolavirus |
|
|
| Ebola virus - Mayinga, Zaire, 1976 | [AF086833=NC_002549] | (EBOV-May) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Ebolavirus but have not been approved as species
| Bundibugyo virus - Bundibugyo, Uganda, 2009 | [FJ217161=NC_014373] | (BDBV-Bun) |
This virus was recently proposed to be the member of a new ebolavirus species (“Bundibugyo ebolavirus”) (Kuhn et al., 2010).
List of unassigned species in the family Filoviridae
None.
List of other related viruses which may be members of the family Filoviridae but have not been approved as species
| Lloviu virus | (LLOV) |
It is expected that this virus will be proposed as the type species (“Lloviu cuevavirus”) of a new genus (“Cuevavirus”) in the family (Kuhn et al., 2010).
Phylogenetic relationships within the family
Phylogenetic relationships are illustrated in Figure 3 on p. 668
Similarity with other taxa
Comparison of filovirus genomes with other mononegaviruses demonstrates a similar structure and suggests comparable mechanisms of transcription and replication. Comparative sequence analyses of single genes indicate that filoviruses are phylogenetically quite distinct from members of other families of the order Mononegavirales. Limited homology exists between the carboxy-terminal part of filovirus GP1,2 and the trans-membrane p15E-related glycoproteins of oncogenic retroviruses.
Derivation of names
Ebola: from the Ebola river in Zaire/Democratic Republic of the Congo, where one of the first registered outbreaks of the disease was thought to have occurred.
Filo: from Latin filum, “thread”, to indicate the morphology of virus particles.
Marburg: from Marburg an der Lahn, a town in Germany, where the first known outbreak of filovirus disease occurred.
*Note
In 2010, the ICTV Filoviridae Study Group and other experts suggested to change the species name Lake Victoria marburgvirus to “Marburg marburgvirus” (Kuhn et al., 2010) and a proposal to this effect is currently under consideration by the ICTV.
Further reading
Klenk , H.-D. (Ed.) ( 1999 ). Marburg and Ebola Viruses: Current Topics in Microbiology and Immunology ( vol. 235 ) . Berlin : Springer-Verlag .
Klenk , H.-D. and Feldmann , H. (Eds.). ( 2004 ). Ebola and Marburg Viruses . Wymondham, Norfolk, UK : Horizon Bioscience .
Kuhn , J.H. ( 2008 ). Filoviruses – a Compendium of 40 Years of Epidemiological, Clinical, and Laboratory Studies . Vienna: Springer Verlag .
Kuhn , J.H. , Becker , S. , Ebihara , H. , Geisbert , T. W. , Johnson , K.M. , Kawaoka , Y. , Lipkin , W.I. , Negredo , A.I. , Netesov , S.V. , Nichol , S.T. , Palacios , G. , Peters , C.J. , Tenorio , A. , Volchkov , V.E. and Jahrling , P.B. ( 2010 ). Proposal for a revised taxonomy of the family Filoviridae : classifi cation, names of taxa and viruses, and virus abbreviations . Arch. Virol. , 155 , 2083 – 2103 .
Martini , G.A. and Siegert , R. (Eds.) ( 1971 ). Marburg Virus Disease . Berlin : Springer-Verlag Pattyn , S.R. (Ed.) ( 1978 ). Ebola Virus Haemorrhagic Fever . Amsterdam : Elsevier/North-Holland Biomedical Press .
Ryabchikova , E.I. and Price , B.B. ( 2004 ). Ebola and Marburg Viruses – A View of Infection Using Electron Microscopy . Columbus, OH : Battelle Press .
Contributed by
Kuhn, J.H., Becker, S., Ebihara, H., Geisbert, T.W., Jahrling, P.B., Kawaoka, Y., Netesov, S.V., Nichol, S.T., Peters, C.J., Volchkov, V.E. and Ksiazek, T.G.
Figures
Figure 1 (Top) Diagram of a filovirion. (Bottom left) Negative contrast electron micrograph of Marburg virus (MARV) particles purified and concentrated by centrifugation from guinea pig serum, 7 days after intraperitoneal infection, and stained with 1% phosphotungstate. (Bottom right) Negative contrast electron micrograph of Ebola virus (EBOV) particles, from Vero cell culture supernatant, 4 days after infection. The bars represent 450 nm.
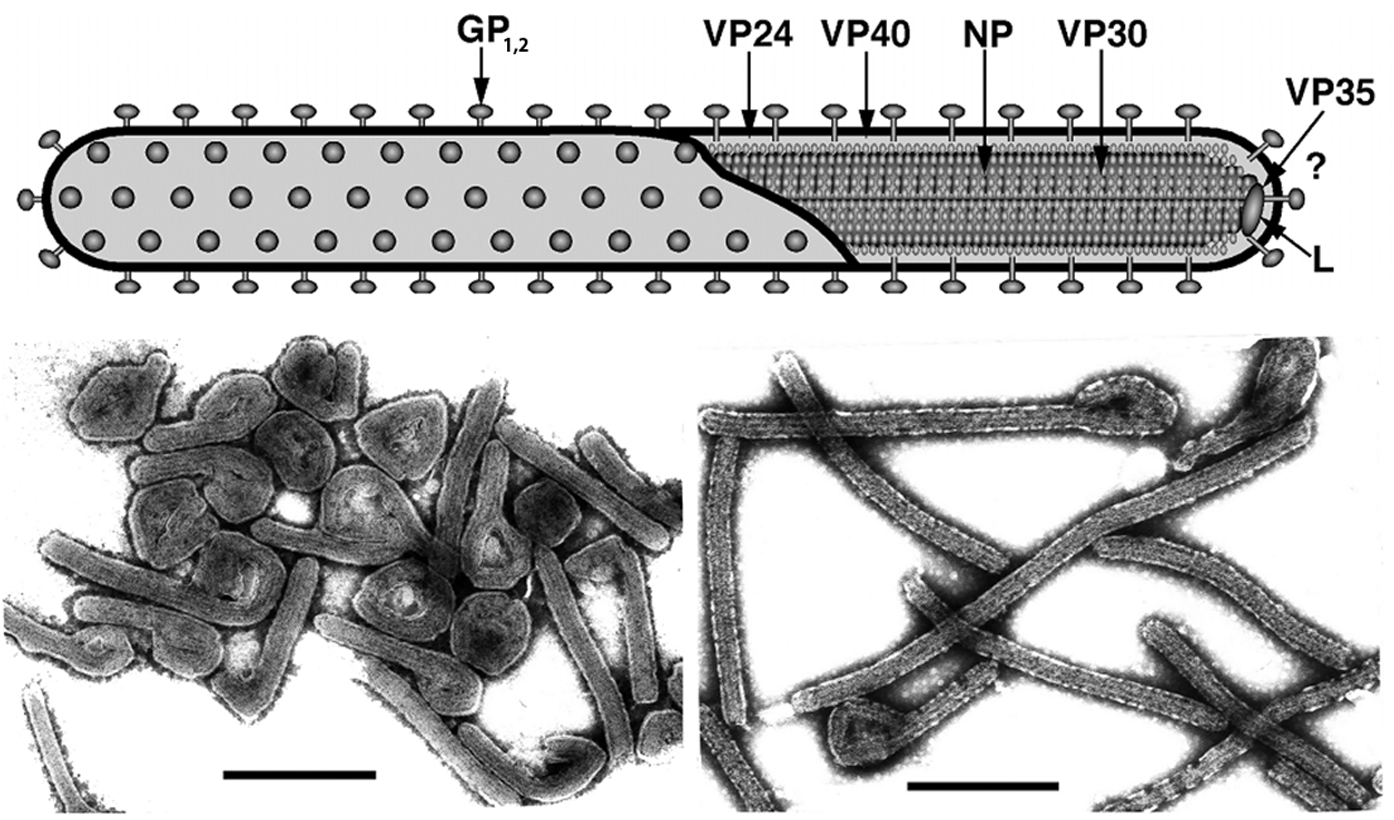
Figure 2 Diagram of filovirus genome organization. The genes that encode structural proteins are identified in the genomes and are drawn to scale. Colored boxes designate coding regions and gray boxes designate non-coding regions. Genes begin with conserved transcriptional initiation sites and end with conserved transcriptional termination sites (polyadenylation sites). Adjoining genes are either separated from one another by an intergenic region (arrows) or overlap. The slippery sequence of the ebolaviral GP gene is indicated by a black triangle. At the extreme 3 and 5 ends of the genomes are leader and trailer sequences, respectively, that are in part complementary.
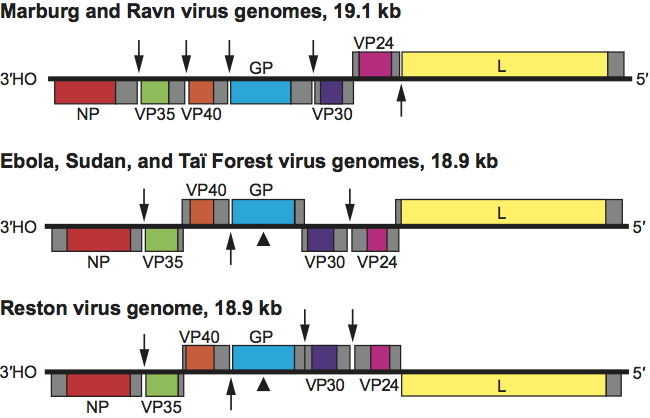
Figure 3 Phylogenetic tree comparing full-length marburgvirus and ebolavirus genomes by Bayesian analysis.