Family: Megabirnaviridae
Yukiyo Sato, Naoyuki Miyazaki, Satoko Kanematsu, Jiatao Xie, Said A. Ghabrial, Bradley I. Hillman and Nobuhiro Suzuki
The citation for this ICTV Report chapter is the summary published as Sato et al., (2019):
ICTV Virus Taxonomy Profile: Megabirnaviridae, Journal of General Virology, 100, 1269–1270.
Corresponding author: Nobuhiro Suzuki ([email protected])
Edited by: Peter Simmonds and Sead Sabanadzovic
Posted: June 2019, updated June 2024
PDF: ICTV_Megabirnaviridae.pdf (2019 version)
This Report chapter is dedicated to the memory of our valued colleague Professor Said Ghabrial.
Summary
Megabirnaviridae is a family of non-enveloped spherical viruses with segmented (2 linear segments, each of 7.2–8.9 kbp) dsRNA genomes, comprising 16.1 kbp in total (Table 1 Megabirnaviridae). There is one genus in the family, Megabirnavirus, to which is assigned the species Rosellinia necatrix megabirnavirus 1 and four other species. The exemplar strain of Megabirnavirus ichi infects the white root rot fungus (Rosellinia necatrix) and confers hypovirulence to the host fungi. Megabirnaviruses are characterized by their bisegmented genome with large untranslated regions (over 1.6 kb) upstream of the 5′-proximal coding strand ORFs, and large protrusions on the particle surface.
Table 1 Megabirnaviridae. Characteristics of members of the family Megabirnaviridae
| Characteristic | Description |
| Typical member | Rosellinia necatrix megabirnavirus 1-W779, (dsRNA1: AB512282; dsRNA2: AB512283), species Megabirnavirus ichi, genus Megabirnavirus |
| Virion | Isometric, non-enveloped particles, 53.5 nm in diameter; dsRNA segments likely to be individually encapsidated |
| Genome | Two linear dsRNAs of 7.2–8.9 kbp, 16.1 kbp in total |
| Replication | No information available |
| Translation | Possible internal ribosomal entry site at the long 5′-untranslated region of over 1.6 kb allows translation of the 5′-proximal open reading frames (ORFs) on the mRNAs from dsRNA1 and dsRNA2. The 3′-proximal ORF of dsRNA1 is translated via −1 ribosomal frameshifting. No information is available the expression of the dsRNA2 downstream |
| Host Range | Fungi |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Duplornaviricota, class Chrymotiviricetes, order Ghabrivirales; one genus and five species |
Virion
Morphology
The genomic dsRNAs of Rossellinia necatrix megabirnavirus 1-W779 (RnMBV1-W779) are encapsidated in non-enveloped spherical particles with 53.5 nm diameter (Figure 1 Megabirnaviridae) (Chiba et al., 2009, Miyazaki et al., 2015, Wang et al., 2023). The major capsid is composed of 60 asymmetrical homodimers (A and B forms) of capsid proteins (CPs) with a T=1 lattice, presumed to produce 120 protrusions on the virus surface, each with a width of ~4.5 nm and a height of ~5.0 nm (Figure 2 Megabirnaviridae) (Miyazaki et al., 2015, Wang et al., 2023). An additional structure termed “crown” is situated over B form MCP at the 3-fold axis. The crown structure consists of a trimer of a truncated protein encoded by the 3’-terminal portion of dsRNA ORF3 (Wang et al., 2023). The major capsid protein (MCP) is encoded by the 5′-proximal ORF on dsRNA1 which also encodes an RNA-directed RNA polymerase (RdRP) by the 3′-proximal ORF (Figure 2 Megabirnaviridae). Purified virions include CP-RdRP fusion protein which is translated from bicistronic transcripts of dsRNA1 (Chiba et al., 2009, Salaipeth et al., 2014).
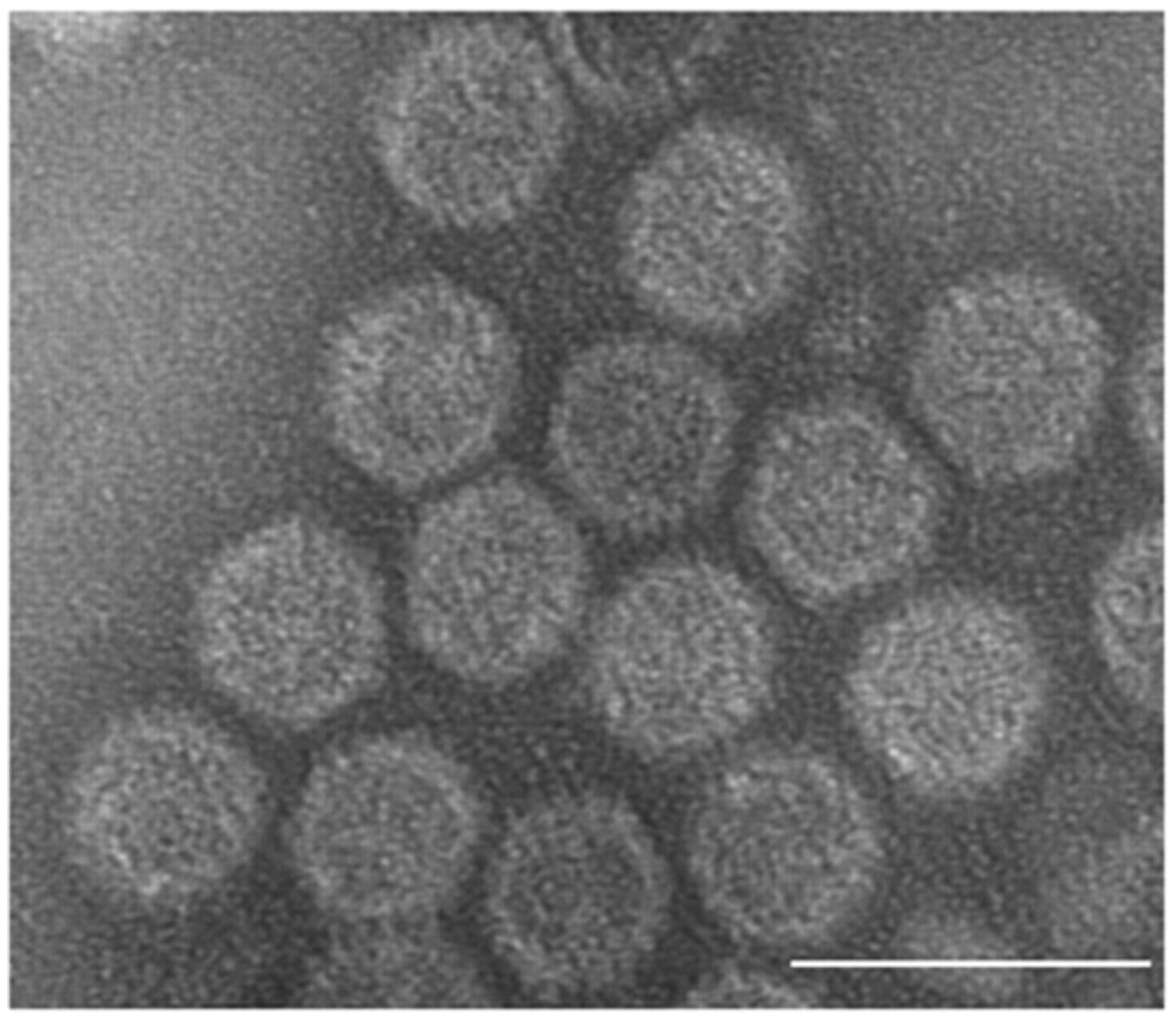 |
| Figure 1 Megabirnaviridae. Transmission electron micrograph of negatively-stained virions of the strain W779 of Rosellinia necatrix megabirnavirus 1 (RnMBV1-W779), a member of the type species of the genus Megabirnavirus. The scale bar represents 100 nm. Reproduced with permission from (Chiba et al., 2009). |
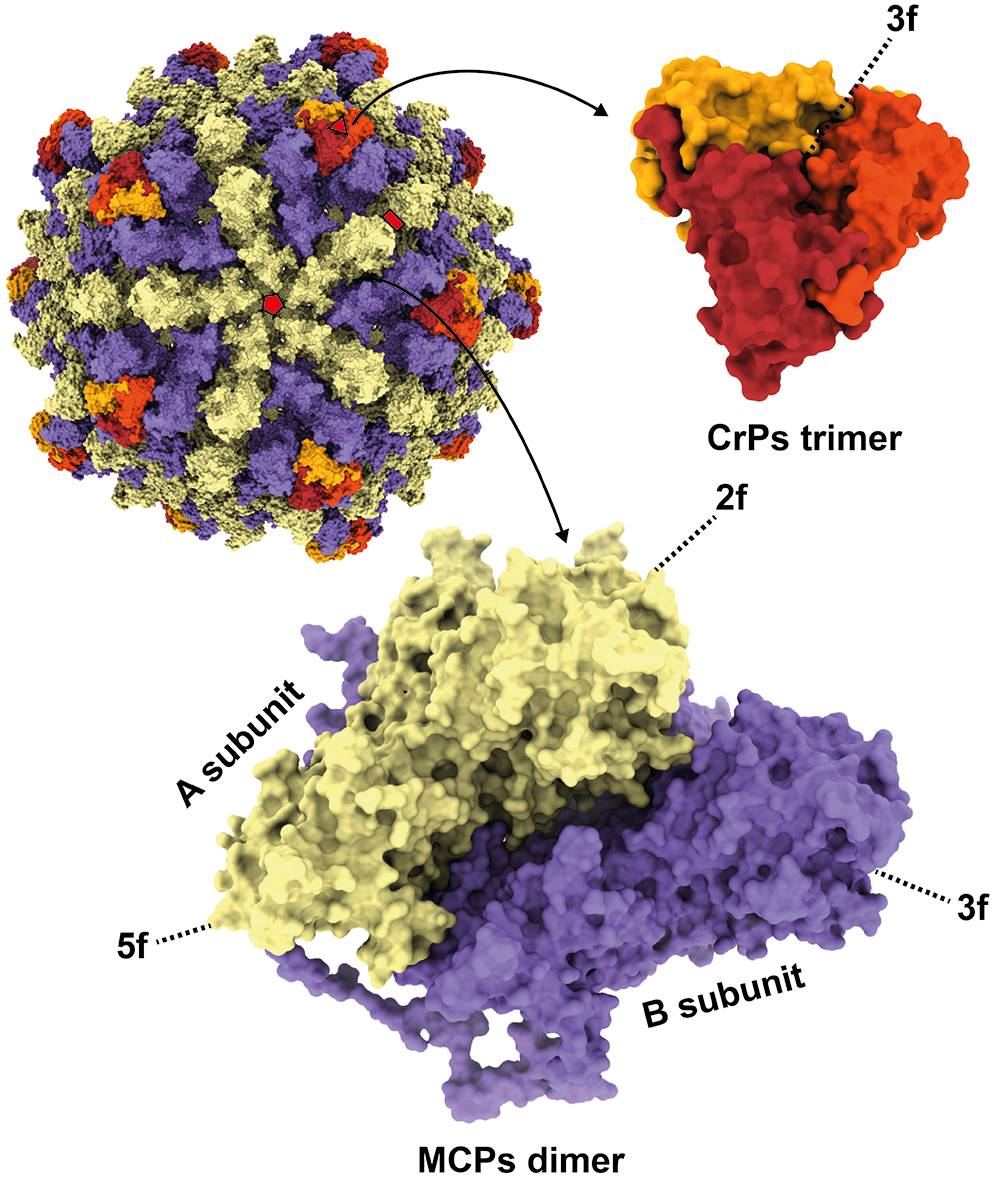 |
| Figure 2 Megabirnaviridae. Three-dimensional cryo-electron microscopy reconstruction of virions of Rosellinia necatrix megabirnavirus 1-W779 (RnMBV1-W779) at a resolution of 3.2 Å. The RnMBV1 capsid is composed of 60 asymmetric homodimers of major capsid protein (MCP) (yellow: MCP_A, purple: MCP_B) and crown protein (CrP) timers (in red, orange, and dark yellow). The red pentagon, triangle, and rectangle show the icosahedral 5-fold (5f), 3-fold (3f), and 2-fold (2f) axes, respectively. Adapted with permission from Wang et al., 2023. |
Physicochemical and physical properties
Unknown.
Nucleic acid
Members of the family Megabirnaviridae possess bipartite linear dsRNA segments, termed dsRNA1 (8.9 kbp) and dsRNA2 (7.2. kbp). There are variable and unequal molar ratios of the two segments in purified virion preparations suggesting that they are packaged separately (Chiba et al., 2009).
Proteins
There is a single major CP and a minor CP-RdRP fusion protein encoded by dsRNA1. Virion-associated RNA polymerase activities have not yet verified. dsRNA2 encodes two non-overlapping ORFs, ORF3 and ORF4. The translated ORF3 product appears to be proteolytically processed into a series of smaller proteins (Kanematsu et al., 2014). No proteins have been detected from expression of the much shorter ORF4.
Genome organization and replication
The genome of RnMBV1-W779 consists of two linear dsRNA segments, each of which contains two open reading frames (ORFs) (Figure 3 Megabirnaviridae) (Chiba et al., 2009). ORF1 and ORF2 on dsRNA1 encode a single major structural protein (CP, P1) and an RNA-directed RNA polymerase (RdRP, P2), respectively. The very C-terminal portion with approximately 135 amino acids of dsRNA2 ORF3-encoded P3 comprises a crown protein structure on the particle surface. The non-overlapping ORF4 on dsRNA2 encodes a hypothetical protein with unknown function. RdRP is encoded in the -1 frame of ORF1 which has a putative slippery sequence (5′-AAAAAAC-3′) at its 3′-terminus followed by a potential stem-loop structure. In accordance with the presence of these signatures of −1 ribosomal frameshifting (Atkins et al., 2016), a CP-RdRP fusion protein is translated from a bicistronic transcript of dsRNA1 and encapsidated in virus particles (Chiba et al., 2009, Salaipeth et al., 2014). P3 accumulates in host cells and is proteolytically cleaved into several smaller proteins (Kanematsu et al., 2014); expression from ORF4 has not been demonstrated (Salaipeth et al., 2014).
The 5′-proximal ORFs in the coding strand of both dsRNA1 and dsRNA2 are preceded by large (over 1.6 kb) untranslated regions which contain multiple mini-ORFs and are likely to direct internal ribosome entry site (IRES)-mediated translation as in the case for other fungal viruses (Chiba et al., 2009, Chiba et al., 2018). The 3′-UTRs of the coding strands of both segments are relatively short (<0.4 kbp). The sequence of both 5′- and 3′-termini is conserved between dsRNA1 and dsRNA2.
Transfection of RnMBV1-W779 virions into fungal cells triggers rearrangement of the virus genome (Kanematsu et al., 2014), namely the loss of dsRNA2 and the emergence of new segments termed as dsRNAS1 composed from the duplicated partial RdRP ORF. The mutant lacking dsRNA2 is competent in replication and packaging, but accumulates lower amounts of viral dsRNAs than wild type. These observations suggest that dsRNA1 is sufficient for RnMBV1 replication as long as it is maintained in laboratory conditions.
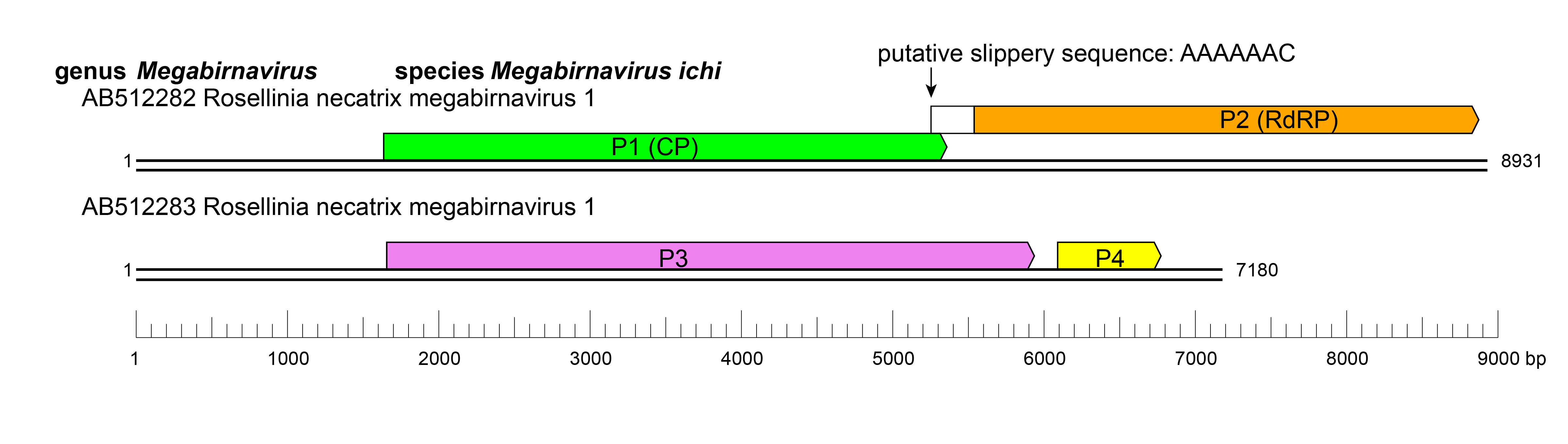 |
| Figure 3 Megabirnaviridae. Genomic organization of Rosellinia necatrix megabirnavirus 1-W779. |
Biology
Megabirnaviruses persistently infect filamentous fungi. The biology of the exemplar isolate Rosellinia necatrix megabirnavirus 1-W779 (RnMBV1-W779) has been well investigated. RnMBV1-W779 is horizontally transmitted between mycelially compatible fungal hosts via hyphal anastomosis and distributed evenly within a single colony (Chiba et al., 2009, Yaegashi et al., 2011). There is little information on extracellular entry and vertical transmission in nature. Purified virions can be introduced into spheroplasts of the natural (Rosellinia necatrix) and the experimental fungal host (Cryphonectria parasitica) (Chiba et al., 2009, Salaipeth et al., 2014). Vertical transmission via asexual sporulation occurs infrequently (<1%) in C. prasitica (Salaipeth et al., 2014).
RnMBV1-W779 inhibits growth of and causes hypovirulence (reduced virulence) of white root rot fungus (R. necatrix) and chestnut blight fungus (C. parasitica), both of which are harmful phytopathogenic fungi (Chiba et al., 2009, Salaipeth et al., 2014). Thus, RnMBV1-W779 is regarded as a potential biological control agent for these pathogens. The molecular mechanism underlying the hypovirulence induced by RnMBV1-W779 remains poorly understood. Mutants of RnMBV1-W779 lacking dsRNA2 but having rearranged dsRNA1 are accumulated less in infected mycelia of R. necatrix and less able to confer hypovirulence on R. necatrix than wild type RnMBV1-W779 (Kanematsu et al., 2014). Thus, dsRNA2 of RnMBV1-W779 is hypothesized to be involved in its hypovirulence and efficient replication. Genome rearrangements also occur following infection of C. parasitica.
RnMBV1-W779 is targetted by antiviral RNA silencing in C. parasitica (Salaipeth et al., 2014, Yaegashi et al., 2016).
Antigenicity
Antibodies against N- and C-terminally truncated recombinant polypeptides derived from CP, RdRP and P3 of RnMBV1-W779 can bind homologous antigens (Salaipeth et al., 2014, Kanematsu et al., 2014). Antibodies against P3 of RnMBV1-W779 can bind multiple proteins specifically in infected mycelia (Salaipeth et al., 2014).
Derivation of names
Megabirnaviridae, Megabirnavirus: from greater genome size (approximately 16 kbp) than those of the members of the family Birnaviridae (approximately 6 kbp) and Picobirnaviridae (approximately 4 kbp), and from bipartite dsRNA genome
Genus demarcation criteria
Not yet defined since only a single genus has been assigned.
Relationships within the family
See discussion under "Related, unclassified viruses" below.
Relationships with other taxa
Members of the family Chrysoviridae, which have relatively close phylogenetic relationships based on their RdRP sequences (Figure 4. Megabirnaviridae), are distinguished from megabirnaviruses by several characteristics such as the number of genome segments (3–7 for chrysoviruses compared with two for megabirnaviruses), virion size (<40 nm in diameter for chrysoviruses compared with 52 nm in diameter for megabirnaviruses) and coding capacity of each segment (mono-cistronic for chrysoviruses but bi-cistronic for megabirnaviruses) (Ghabrial et al., 2018, Ghabrial et al., 2018, Sato et al., 2018).
 |
| Figure 4 Megabirnaviridae. Phylogenetic analysis based on the complete deduced amino acid sequences of RdRPs of the member of the family Megabirnaviridae, related, unclassified viruses, and phlegiviruses, chrysoviruses, totiviruses and botybirnaviruses. The sequences were aligned using MUSCLE (Edgar 2004) integrated in MEGA7 (Kumar et al., 2016). The evolutionary history was also inferred using the Neighbor-Joining method and the evolutionary distances were computed using the Poisson correction method in MEGA7. The percentage of reproducibility of trees in the bootstrap test (1000 replicates) is shown next to the branches of the midpoint-rooted tree. Viruses that have not been formally assigned to the family Megabirnaviridae are indicated by a grey bar. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Members of the family Orthototiviridae and the genus Botybirnavirus also have distant phylogenetic relationships with the megabirnaviruses (Figure 4 Megabirnaviridae). Totiviruses have a non-segmented bicistronic dsRNA genome. The RdRP of fungal totiviruses (members of the genera Totivirus and Victorivirus) is encoded downstream of the CP gene, resembling the genome arrangement of dsRNA1 of megabirnaviruses. Totiviruses with undivided dsRNA genomes are phylogenetically distinct from megabirnaviruses (Figure 4 Megabirnaviridae) and have coding strands with a shorter 5′-UTR (<0.5 kb) and smaller particles (approximately 40 nm in diameter).
Members of the family Partitiviridae and the genus Botybirnavirus also have bipartite dsRNA genomes. Partitiviruses and botybirnaviruses have relatively small, monocistronic genomes (approximately 4 kbp for partitiviruses and approximately 12 kbp for botybirnaviruses) and smaller virions (approximately <40 nm in diameter) compared to megabirnaviruses (Sato et al., 2018). Partitiviruses show more distant phylogenetic relationships to megabirnaviruses than to chrysoviruses and totiviruses (Chiba et al., 2009, Vainio et al., 2018).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Number and size of genome segments (bp) |
| Entoleuca megabirnavirus 1-97-14 | RNA1: MF375886 | EnMV1-97-14 | 1? (8,927) |
| Rosellinia necatrix megabirnavirus 3-Rn454 | RNA1: LC333756 | RnMBV3-Rn454 | 1? (8,967) |
| Rhizoctonia solani megabirnavirus 1-DC17 | KX349071a | RsMBV1-DC17 | 1? |
| Rhizoctonia solani RNA virus-HN008 | RNA1: KP861921 | RsRV-HN008 | 1 (7,596) |
Virus names and virus abbreviations are not official ICTV designations.
a Only partial RdRP sequence available.
The family Megabirnaviridae includes the single genus Megabirnavirus to which the species Megabirnavirus ichi and four other species have been assigned. Database searching and phylogenetic analysis based on complete or partial RdRP amino acid sequences reveals several unclassified viruses closely related to the member of this species (Table 2 Megabirnaviridae, Figure 4 Megabirnaviridae). Similarities are also seen in the P1 and P3 sequences of these megabirna-related viruses (Table 2 Megabirnaviridae).
Table 2 Megabirnaviridae. Identity of amino acid sequence of hypothetical proteins of unclassified viruses related to Rosellinia necatrix megabirnavirus 1-W779 of the family Megabirnaviridae.
| Virus name | RdRPa | P1 (CP)a | P3a | Reference |
| Sclerotinia sclerotiorum megabirnavirus 1-SX466 | 44% | 36% | 33% | (Wang et al., 2015) |
| Rosellinia necatrix megabirnavirus 2-W8 | 47% | 35% | 54% | (Sasaki et al., 2016) |
| Pleosporales megabirnavirus 1-MUT4917 | 44% | 27% | 24% | (Nerva et al., 2016) |
| Entoleuca megabirnavirus 1-97-14 | 38% | 19% | c | (Velasco et al., 2019) |
| Rosellinia necatrix megabirnavirus 3-Rn454 | 38% | 22% | c | (Arjona-Lopez et al., 2018) |
| Rhizoctonia solani megabirnavirus 1-DC17 | 39%d | b | c | (Bartholomäus et al., 2016) |
| Rhizoctonia solani RNA virus-HN008 | 30% | n.d. b | Absent | (Zhong et al., 2015) |
a Percentage amino acid identity with RnMBV1-W779 estimated by protein BLAST. No virus element which shows substantial identity to P4 of RnMBV1-W779 was found.
b No sequence similarity detected.
c Complete sequence unavailable.
These megabirnavirus-related viruses infect filamentous ascomycetes and basidiomycetes. Complete genome sequences are available for four viruses i.e., Sclerotinia sclerotiorum megabirnavirus 1 (SsMBV1), Rosellinia necatrix megabirnavirus 2 (RnMBV2), Pleosporales megabirnavirus 1 (PMbV1) and Fusarium pseudograminearum megabirnavirus 1 (FpgMBV1), and these have close phylogenetic relationships with RnMBV1 (Wang et al., 2015, Sasaki et al., 2016, Nerva et al., 2016, Zhang et al., 2018). These four viruses and RnMBV1 have many common characteristics including: (1) genomes consist of bipartite segments (RNA1: 8.8–9.0 kbp; RNA2: 5.1–8.0 kbp) with a very long UTR (1.1–1.8 kb) upstream of 5′-proximal coding strand ORFs; (2) coding sequences for CP and RdRP are on the larger segment, dsRNA1; (3) there are two ORFs encoding P3 and P4 on dsRNA2. There is little or no sequence similarity among P4s of related megabirnaviruses. SsMBV1, RnMBV2 and FpgMBV1 were isolated from plant-pathogenic fungi, while PMbV1 was isolated from marine fungi associated with seagrass. The dsRNA2 of PMbV1 and FpgMBV1 (5.1–5.3 kbp) is shorter than dsRNA2 of RnMBV1, SsMBV1 and RnMBV2 (7.2–8.0 kbp). The putative slippery sequence at the 3′-terminus of the CP ORF (ORF1) is highly conserved in RnMBV1, SsMBV1 and RnMBV2, but differs from that found in PMbV1 and FpgMBV1.
The P3 protein encoded by SsMBV1 dsRNA2 contains a papain-like protease domain which appears to have been horizontally transferred from a positive-sense (+) single-stranded RNA virus (hypovirus) (Wang et al., 2015). This horizontal gene transfer between a dsRNA and a (+)ssRNA virus is a rare event. Such a papain-like protease domain is not found in P3 of RnMBV1 and RnMBV2, although the deduced amino acid sequence of P3 of these three viruses all show moderate levels of sequence identity (Table 2. Megabirnaviridae). RnMBV2 shares a common fungal host, R. necatrix, with RnMBV1. Single infection of RnMBV2, however, causes no obvious phenotypic change of R. necatrix in contrast to RnMBV1-W779 (Sasaki et al., 2016), while double infection with RnPV1 induces hypovirulence. Notably, RnMBV2 can survive a complete loss of dsRNA2 under the laboratory conditions (Sasaki et al., 2016). SsMBV1 slightly inhibits the growth of host fungus.
Three partially sequenced viruses, namely Rizoctonia solani megabirnavirus 1 (RsMBV1), Rosellinia necatrix megabirnavirus 3 (RnMBV3) and Entoleuca megabinavirus 1 (EnMV1), are also closely related to RnMBV1 (Figure 4. Megabirnaviridae) (Arjona-Lopez et al., 2018, Bartholomäus et al., 2016) (Velasco et al., 2019). Information on dsRNA2 in these viruses is unavailable. A long UTR (>1.6 kb) is present upstream of the most 5′-proximal of the two ORFs on the coding strand of dsRNA1 of RnMBV3 and EnMV1, similar to dsRNA1 of RnMBV1.
Rhizoctonia solani RNA virus HN008 (RsRV-HN008) is also placed in the same phylogenetic clade as RnMBV1 (Figure 4 Megabirnaviridae) (Zhong et al., 2015). RsRV-HN008 lacks dsRNA2 and has only one genome segment with two in-frame non-overlapping ORFs. Protein encoded by ORF1 of RsRV-HN008 shows no sequence similarity to CP of megabirnaviruses. The UTR upstream of the 5′-proximal ORF on the coding strand of the genome segment of RsRV-HN008 is much shorter (39 nt) than that of megabirnaviruses. Thus, RsRV-HN008 seems peculiar in this phylogenetic clade and may represent an intermediate between megabirnaviruses and phlegiviruses (see below).
The unclassified viruses within the proposed genus phlegivirus are closely related to megabirnaviruses (Figure 4 Megabirnaviridae) (Petrzik et al., 2016). Phlegiviruses have non-segmented dsRNA genomes with two ORFs. ORF2 of phlegiviruses is positioned in the -1 frame relative to ORF1. A putative slippery sequence immediately upstream of the stop codon of ORF1 suggests −1 ribosomal frameshifting for translation of the downstream ORF2. Phlegivirus ORF1 encodes hypothetical protein containing the NUDIX domain, and shows no significant similarity to megabirnavirus CP. Some phlegiviruses such as Rhizoctonia fumigata virus 1, Lentinula edodes mycovirus (LeV)-HKB and Phlebiopsis gigantea large virus 1 have relatively long UTRs (0.5–0.9 kb) upstream of the 5′-proximal ORF on the coding strand, while others such as Thelephora terrestris virus 1 do not (Petrzik et al., 2016, Magae 2012, Lim et al., 2011, Li et al., 2015). No phlegiviruses particles have been observed except for spherical particles of Lentinula edodes spherical virus which is regarded as an isolate of LeV (Won et al., 2013). Further study is required for classification of these viruses.
The unclassified viruses of the proposed family fusagraviridae also have non-segmented dsRNA genome with two non-overlapping ORFs (Wang et al., 2016). Fusagraviruses show more distant phylogenetic relationships to megabirnaviruses than to chrysoviruses and totiviruses (Wang et al., 2016). Fusagravirus ORF1 encodes a hypothetical protein with unknown function, which is not related to CPs of known mycoviruses. Fusagravirus ORF2 encodes a putative RdRP which includes a conserved Phytoreo_S7 domain and is hypothesized to be translated by −1 ribosomal frameshifting based on the presence of a slippage signature sequence. The UTR upstream of the 5′-proximal ORF on the coding strand of fusagraviruses is long (0.87–1.31 kb), like that of megabirnaviruses. It remains an open question whether fusagraviruses are encapsidated. Fusagraviruses can be distinguished from megabirnaviruses in the number of genome segments and in their phylogenetic placement.

