Family: Adenoviridae
Genus: Barthadenovirus
Distinguishing features
Barthadenoviruses are serologically distinct from viruses in the other adenovirus genera, and their genomic organization and capsid protein complements also differ (Vrati et al., 1996). The peculiar biological properties (growing only on primary testicle cells instead of established cell lines, etc.) of some bovine adenovirus types led Adorján Bartha, a Hungarian veterinary virologist, to recognise that they represent a specific adenovirus lineage (Bartha 1969). Later molecular and phylogenetic analyses proved the existence of a separate evolutionary lineage also involving avian, reptilian and marsupial hosts and led to the proposal of a new genus (Harrach et al., 1997, Benkő and Harrach 1998, Benkő et al., 2002, Farkas et al., 2002). Barthadenoviruses have been detected in a broad range of hosts, predominantly including scaled reptiles such as snakes (Benkő et al., 2002, Garner et al., 2008, Abbas et al., 2011, Bogan et al., 2024a, Bogan et al., 2024b), lizards (Wellehan et al., 2004, Papp et al., 2009, Ball et al., 2014, Prado-Irwin et al., 2018, Hyndman et al., 2019), and worm lizards (Szirovicza et al., 2016), but also birds (Harrach et al., 1997, Hess et al., 1997, To et al., 2014, Duarte et al., 2019, Needle et al., 2019b, Athukorala et al., 2020, Shan et al., 2022) and ruminants (Vrati et al., 1996, Harrach et al., 1997, Dán et al., 1998, Fox et al., 2017, Miller et al., 2017, Woods et al., 2018, Paim et al., 2021). There have been sporadic reports of their occurrence in marsupials (Thomson et al., 2002, Gál et al., 2017, Okoh et al., 2023a) and non-squamate reptiles (tortoises) (Garcia-Morante et al., 2016, Salzmann et al., 2021, Bogan et al., 2024b).
Virion
Morphology
Barthadenovirus particles are largely similar to those of mastadenoviruses, except for the prominent knobs formed by protein LH3 at the icosahedral and local threefold axes in the facet (Menéndez-Conejero et al., 2017). This distinguishes barthadenoviruses structurally from other known adenoviruses (Figure 1 Barthadenovirus). LH3 is located in the same relative position among the hexon subunits as protein IX (present only in mastadenoviruses) but sits on top of the hexon towers, rather than in the valley between hexon bases (Liu et al., 2010, Menéndez-Conejero et al., 2017). Lizard adenovirus 2 (LAdV-2) LH3 contacts the capsid surface via a triskelion structure identical to that used by mastadenovirus protein IX (Marabini et al., 2021). The high resolution structure of LAdV-2 shows a large conformational difference in the internal vertex protein IIIa between mast- and barthadenoviruses, possibly related to the presence of an extended polypeptide absent in HAdVs (Marabini et al., 2021). This polypeptide and α-helical clusters beneath the facet are proposed to correspond to genus-specific proteins LH2 and p32K. Ovine adenovirus 7 (OAdV-7) particles that lack the LH3 or p32K proteins are viable although less stable (Pantelic et al., 2008). Similarly, psittacine adenovirus 3 (PsAdV-3) and white-eyed parakeet adenovirus 1 (GenBank accession no. KJ675568, mislabelled as PsAdV-3), both types belonging to species Barthadenovirus amazonae, and psittacine adenovirus 11 (PsAdV-11, from blue-throated macaw, Barthadenovirus caerulei) seem to lack LH3 (To et al., 2014, Duarte et al., 2019, Shan et al., 2022). (All non-mastadenoviruses lack proteins V and IX but are nonetheless viable.)
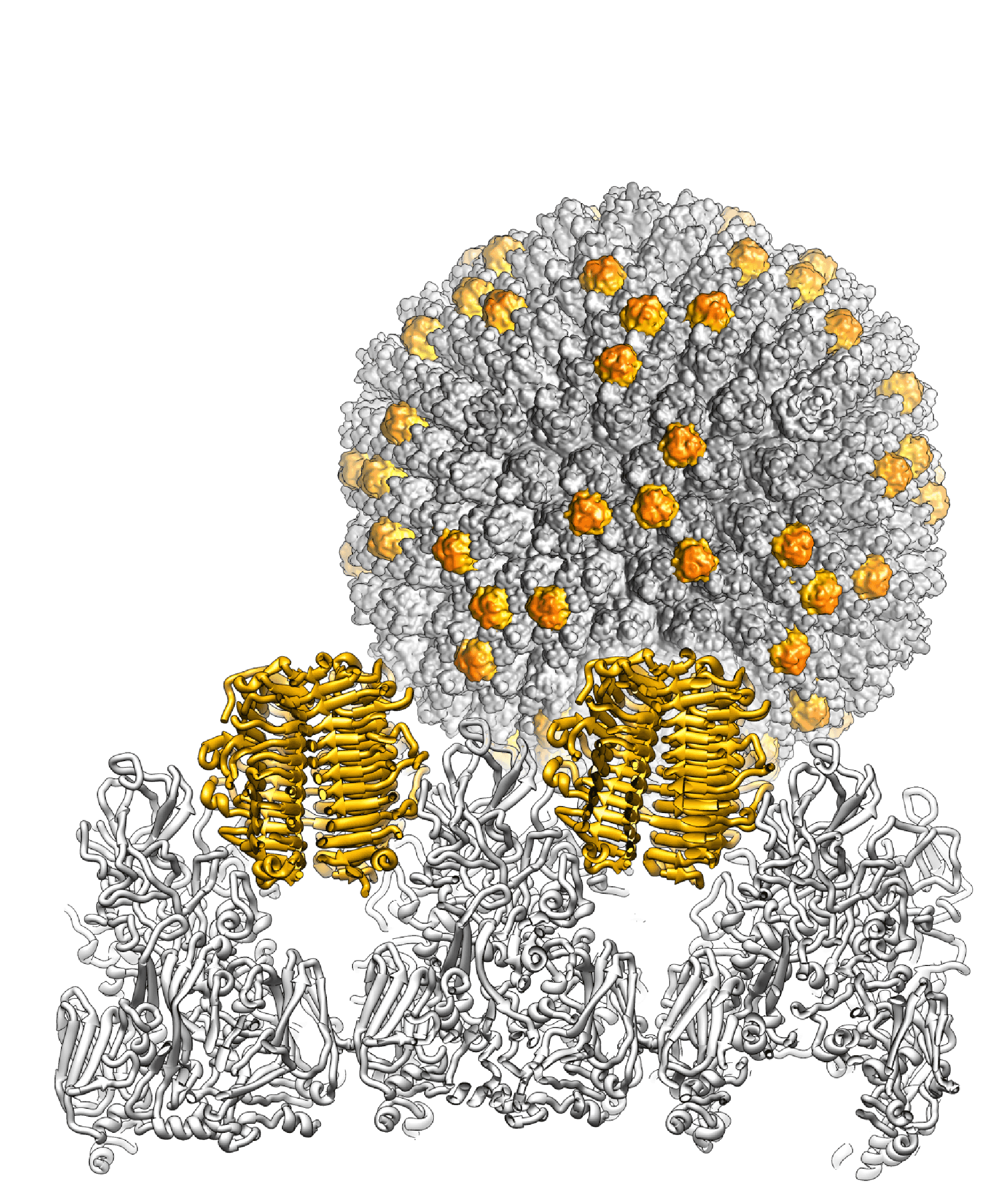 |
| Figure 1 Barthadenovirus Cryo-EM map of the snake adenovirus 1 virion at 11 Å resolution is shown in the background, with the genus-specific minor coat protein LH3 highlighted in yellow. In the foreground, the crystal structure of LH3 (yellow) and its position with respect to hexons (grey) is shown in a view across the particle. LH3 has a fold typical of bacteriophage tailspikes, and makes multiple contacts with hexons to stabilize the capsid (Menéndez-Conejero et al., 2017). |
Lizard adenovirus 2 (from Mexican beaded lizard, Barthadenovirus lacertae) has three long fibers in some vertices and single short fibers in the rest (Pénzes et al., 2014). The 3D structures of snake adenovirus 1 (SnAdV-1, Barthadenovirus serpentis) (Singh et al., 2014) and bovine adenovirus 4 (Barthadenovirus quartum) fibers have been established (Nguyen et al., 2015).
Physicochemical and physical properties
Virions possess elevated heat stability and retain substantial infectivity after treatment for 30 min at 56°C (Bartha and Kisary 1970). These conditions inactivate mastadenovirus particles.
Nucleic acid
The genome size of sequenced isolates ranges from 27,751 (SnAdV-1) (Farkas et al., 2008) to 39,644 bp (eastern spinebill adenovirus, “passerine adenovirus 1”) (Athukorala et al., 2020) with inverted terminal repeats (ITRs) of 36 bp (bovine adenovirus 7 and scaly thrush adenovirus 1 [Zoothera dauma]) (Dán et al., 2001, Kumagai et al., 2021, Zheng et al., 2024) to 195 bp (bearded dragon adenovirus 1, BDAdV-1) ((Dán et al., 2001, Pénzes et al., 2020b, Kumagai et al., 2021). For ruminant barthadenoviruses, duck adenovirus 1 (DAdV-1), common tern adenovirus 1, grey warbler adenovirus 1 and agile wallaby AdV-1, the G+C composition of the DNA is low, varying between 33.2 (deer adenovirus 1 [OdAdV-1]), and 43.0% (DAdV-1). However, it turned out that barthadenoviruses originating from scaled reptiles have a non-biased nucleotide composition (50.2% G+C in SnAdV-1) (Farkas et al., 2008), 44.15% in LAdV-2 (Pénzes et al., 2014), 45.1% in viviparous lizard adenovirus 1, 55.6% in spiny-tailed monitor adenovirus 1 and 56.3% in BDAdV-1. Additional newly identified avian barthadenoviruses also have a non-biased nucleotide composition: 44.1–53.7% G+C in grey warbler adenovirus 1 (French et al., 2023), white-eyed parakeet adenovirus 1 (Duarte et al., 2019), PsAdV-3 (To et al., 2014) and eastern spinebill adenovirus 1 (“passerine adenovirus 1”) (Athukorala et al., 2020).
Proteins
The proteins of a typical barthadenovirus are summarized in Table 1 Barthadenovirus (Vrati et al., 1996). Barthadenoviruses lack proteins V and IX but have a unique structural protein p32K. LH2 has also been found in purified barthadenovirus particles (SnAdV-1), and is therefore another genus-specific structural protein (Menéndez-Conejero et al., 2017). Table 1 Barthadenovirus. Proteins encoded by ovine adenovirus 7
| kDa | Transcription class | Description | Note |
|---|---|---|---|
| 32 | Unknown | S; p32K† | Unique to barthadenoviruses |
| 13 | LH1 | R | Missing in all avian barthadenoviruses, and agile wallaby AdV-1 |
| 14.7 | LH2 | S | |
| 42.8 | LH3Unknown | R, S | Distant homologue of mastadenovirus E1B 55K, missing in PsAdV-3, white-eyed parakeet AdV-1 and PsAdV-11 |
| 43 | E2 | D; DBP | |
| 123 | E2 | D; pol | |
| 67.1 | E2 | D, S; pTP† | |
| 12.9, 20.9, 19.8, 19.8 | RH1, RH2, RH4, RH6, early | R | F-box proteins unique for barthadenoviruses.Related to each other; missing in PsAdV-3. |
| 22.6 | RH5, early | R | Missing in DAdV-1, PsAdV-3, SnAdV-1, LAdV-2 and BDAdV-1 |
| 17.1 | E4.1, early | R | Missing in DAdV-1 |
| 25.6, 30.8 | E4.2, E4.3, early | R | Distant homologues of mastadenovirus E4 34K |
| 38.2 | Early and late | D; 52/55 kDa*,† | |
| 58.4 | Late | S (pIIIa);† p-protein | |
| 51 | Late | S (III); penton base* | Lacks integrin-binding motif |
| 12.9 | Late | S (pVII);† major core | |
| 7.3 | Late | S (pX);† X/µ | |
| 24.5 | Late | S (pVI)† | |
| 102 | Late | S (II); hexon | |
| 23 | Late | D, S; protease | |
| 72 | Late | D; 100 kDa* | Hexon assembly protein |
| 15.7 | Late | D, R; 33 kDa* | p-protein not found in DAdV-1 |
| 24.7 | Late | S (pVIII)† | |
| 58.2 | Late | S (IV); fiber | Cell attachment protein. Fibers of two lengths in LAdV-2, PsAdV-3 and white-eyed parakeet AdV-1 (both psittacine AdVs belong to B. amazonae). |
| 37.5 | Unknown | D, S (IVa2); packaging |
Molecular masses are presented as unmodified and uncleaved gene products. D = DNA synthesis and packaging; DBP = DNA-binding protein; LH = left end [genes]; p = precursor; p-protein = phosphoprotein; pol = DNA polymerase; R = regulation, RH = right end [genes]; S = structural; TP = terminal protein; All non-structural proteins are hypothetical until characterized. * = Mr values are significantly different from those obtained by SDS-PAGE; † = cleaved by viral protease.
Lipids
None reported.
Carbohydrates
See discussion under family properties.
Genome organization and replication
The central part of the barthadenovirus genome is similar to that of mastadenoviruses (except that there are no protein V and IX genes), whereas the proteins encoded by the extremities of the genomes differ markedly (Vrati et al., 1996, Hess et al., 1997, Farkas et al., 2008, Pénzes et al., 2014, To et al., 2014, Miller et al., 2017, Pénzes et al., 2020b, Kumagai et al., 2021, Matsvay et al., 2021, Zheng et al., 2024). The left end of the genome carries a gene for p32K, a unique structural protein. At this end, gene LH1 is also unique to the genus but is not present in all members (it seems to be missing in all the avian barthadenoviruses) (Kraberger et al., 2022). LH2 is another genus-specific gene (Menéndez-Conejero et al., 2017). The right end of the genome contains genes that are related to each other, suggestive of gene duplication. There are two E4 34K gene homologs (E4.2 and E4.3), and one to seven RH gene homologues. At this end of the genome, genes E4.1 and RH0–RH6 are also unique to the genus but are not present in all members. The proteins encoded by genes LH3 and E4.3 (and its paralogue, E4.2) show limited similarity to mastadenovirus proteins E1B 55K and E4 34K, respectively, and (attached to each other) have been proposed to play a similar role, i.e. targeting cellular proteins for degradation (Gilson et al., 2016).
No immunomodulatory genes such as those present in the mastadenovirus E3 region have yet been identified. DAdV-1 and PsAdV-3 have a unique region at the far-right end of the genome that contains seven and six uncharacterized ORFs, respectively. Six and five of these are different in these two barthadenoviruses, respectively, and may be avian host-specific, whereas both viruses have ORF1, which is shared by LAdV-2 and BDAdV-1. Both of these reptile barthadenoviruses have also five further genes in this region; these are not homologous with those in the other lizard adenoviruses (Pénzes et al., 2014, Pénzes et al., 2020b). ORF4 and ORF5 of PsAdV-3 are related to each other and also to ORF2 of the aviadenoviruses psittacine adenovirus 1 (PsAdV-1) and psittacine adenovirus 4 (PsAdV-4), and ORF52 of the aviadenoviruses pigeon adenovirus 1 (PiAdV-1) and pigeon adenovirus 2. In these four aviadenovirus types (the two psittacine and two pigeon AdVs), these four homologous genes are located at the far-left end of the genome instead of the right end as they are in the barthadenoviruses. This unique region of DAdV-1 also contains a VA RNA gene that may be homologous to that of the aviadenovirus fowl adenovirus 1 (FAdV-1). The BDAdV-1 genome harbours three genes in this region that encode proteins of the C-type lectin-like domain superfamily (Pénzes et al., 2020b). The protein named ORF3 in BDAdV-1 has a CTLD group II-like domain architecture displaying structural similarity to natural killer cell surface receptors and to an alphaherpesvirus virulence factor for neurotropism, UL45.
The receptor-binding properties of DAdV-1 were predicted from the crystal structure of the fiber head to involve binding to the chicken coxsackievirus and adenovirus receptor (CAR) (Song et al., 2019). Splicing is presumed in the IVa2, pTP and 33K genes but not in the pol gene of barthadenoviruses except BDAdV-1 (Pénzes et al., 2020b).
Biology
Certain barthadenoviruses can cause haemorrhagic epizootic disease in free-living ruminants (Fox et al., 2017, Miller et al., 2017, Woods et al., 2018). DAdV-1 is also associated with a specific disease of hens that is characterized globally by sharp decreases in egg production (egg drop syndrome) and eggshell deformations (Hess et al., 1997). BDAdV-1 has been linked to sudden death, lethargy, weakness, diarrhoea, dehydration and anorexia, and is responsible for central nervous system signs observed in young bearded dragons (“star gazing”, head tilt, opisthotonos, paresis and circling) (Pénzes et al., 2020b).
Due to the lack of pre-existing immunity in humans and its low bio-safety profile, OAdV-7 has been developed as a gene delivery vector intended for human vaccine and gene therapy applications (Both 2004).
Antigenicity
See discussion under family properties.
Species demarcation criteria
Species designation depends on at least two of the following characteristics:
- Phylogenetic distance (>10–15%, based on maximum likelihood analysis of the pol amino acid sequence)
- Host range
- Nucleotide composition
- Cross-neutralization
- Gene organization at the right end of the genome
Related, unclassified viruses
| Virus name | Accession number | Abbreviation |
| Aldabra giant tortoise adenovirus | OR062096 | |
| amphisbaenian adenovirus 1 | KT932964 | |
| anolis adenovirus 1 | KC544015 | |
| anolis adenovirus 2 | KC544016 | |
| anolis adenovirus 3 | KF886534 | |
| Australian ibis adenovirus | MN238659 | |
| blue-tongued skink adenovirus | AY576682 | |
| caiman lizard adenovirus | PQ861825 | |
| chameleon adenovirus 1 | AY576679 | ChAdV-1 |
| chameleon adenovirus 2 | KF886533 | ChAdV-2 |
| chimney swift adenovirus 1 | MG736957 | CSAdV-1 |
| common chaffinch adenovirus | MN380559 | |
| common starling adenovirus | MN238657 | |
| Darwin's finch adenovirus 1 | PV739274 | |
| dunlin (Charadriiformes) adenovirus | PP319130 | |
| emerald monitor adenovirus | EU914208 | |
| eastern box turtle adenovirus 3 | PQ066476 | |
| eastern box turtle adenovirus 4 | PQ066482 | |
| Eurasian bullfinch adenovirus | MN380558 | |
| Eurasian nuthatch adenovirus | OL603901 | |
| Eurasian siskin adenovirus | MN380560 | |
| Eurasian tree sparrow adenovirus | OL603903 | |
| European greenfinch adenovirus strain 47975 | MN380551 | |
| European robin adenovirus | MN380552 | |
| falcated duck adenovirus | PP319137 | |
| gecko adenovirus 1 | AY576677 | GeAdV-1 |
| Greek (spur-thighed) tortoise adenovirus 1 | KT310086 | |
| great cormorant adenovirus 2 | PP319098 | |
| Japalura tree dragon adenovirus 1 | KF886532 | |
| kowari adenovirus | KT696557 | |
| lacertid adenovirus 1 | KT950888 | |
| lacertid adenovirus 2 | KT950886 | |
| little wattlebird adenovirus | MN238660 | |
| lizard adenovirus 1 (gila adenovirus 1) | AY576680 | LAdV-1 |
| long-billed corella adenovirus | MN238656 | |
| long-tailed finch adenovirus | MK413651 | |
| long-tailed grass lizard adenovirus 1 | KM026520 | |
| Mojave rattlesnake adenovirus 1 | PQ066468 | |
| noisy miner adenovirus | MN238663 | |
| pine grosbeak adenovirus (gbk055ade1nc) | MT138097 | |
| psittacine adenovirus 9 (cockatiel) | OK058273 | PsAdV-9 |
| psittacine adenovirus 10 (rose-ringed parakeet) | OK058274 | PsAdV-10 |
| rattlesnake adenovirus 4 | PQ066483 | |
| Russian (Horsfield’s) tortoise adenovirus | MT900853 | |
| satin bowerbird adenovirus | MT079819 | |
| short-tailed pygmy chameleon adenovirus 1 | KM026523 | |
| silver gull adenovirus | MN238658 | |
| silvereye (Passeriformes) adenovirus | MN238665 | |
| snake adenovirus 2 | FJ012163 | SnAdV-2 |
| snake adenovirus 3 | FJ012164 | SnAdV-3 |
| song thrush adenovirus | MN380548 | |
| spotted redshank (Charadriiformes) adenovirus | PP319141 | |
| sulphur-crested cockatoo adenovirus | MN238655 | |
| superb fairywren adenovirus | MN238664 | |
| tokay gecko adenovirus | AY576681 | |
| tropical screech owl adenovirus 1 | MN540447 | |
| vitelline masked weaver adenovirus strain 37869 | MN380538 | |
| vitelline masked weaver adenovirus strain 38132 | MN380539 | |
| white plumed honeyeater adenovirus 1 | MN238667 | |
| white plumed honeyeater adenovirus 2 | MN238668 | |
| white-throated monitor adenovirus 1 | KM026519 | |
| yellow warbler adenovirus ("Passerine barthadenovirus") | PV739278 |
Virus names and virus abbreviations are not official ICTV designations.
Not all adenovirus names have generally accepted and utilised abbreviations. Many sequences have been gained only by PCR from random bird samples (Rinder et al., 2020, Zadravec et al., 2022, Harrach et al., 2023).

