Family: Guttaviridae
David Prangishvili, Tomohiro Mochizuki and Mart Krupovic
The citation for this ICTV Report chapter is the summary published as Prangishvili et al., (2018):
ICTV Virus Taxonomy Profile: Guttaviridae, Journal of General Virology, 99: 290–291.
Corresponding authors: David Prangishvili ([email protected]) and Mart Krupovic ([email protected])
Edited by: Andrew M. Kropinski and Peter Simmonds
Posted: February 2018, updated March 2022
PDF: ICTV_Guttaviridae.pdf (2018 version)
Summary
Guttaviridae is a family of enveloped viruses infecting hyperthermophilic archaea (Table 1.Guttaviridae). The virions are ovoid or droplet-shaped, with a diameter of 55–80 nm and a length of 75–130 nm. The genome is a circular dsDNA molecule of around 14 kbp. The family includes one genus, Betaguttavirus, with a single species.
Table 1.Guttaviridae. Characteristics of members of the family Guttaviridae.
| Characteristic | Description |
| Example | Aeropyrum pernix ovoid virus 1 (HE580237), species Betaguttavirus kodakarajimaense |
| Virion | Enveloped virions of ovoid shape, with a diameter of 55 nm and a length of 75 nm. |
| Genome | Circular dsDNA of about 14 kbp |
| Replication | Genome is likely to be replicated by the host replisome |
| Translation | Not known |
| Host range | Hyperthermophilic archaea, phylum Crenarchaeota |
| Taxonomy | One genus: Betaguttavirus |
Virion
Morphology
The virions of members of the family Guttaviridae have an ovoid shape, measuring 55 × 75 nm (for Aeropyrum pernix ovoid virus 1 (APOV1), Betaguttavirus) when analysed by cryo-electron microscopy (Mochizuki et al., 2011). The virion surface is covered by globular subunits, which are about 3.5 nm in width. In negative-contrast electron micrographs, the virions are slightly pleomorphic, most displaying a droplet-like shape (Figure 1 Guttaviridae). The droplet-shaped morphology is unprecedented among viruses of bacteria and eukaryotes, and represents a group of archaea-specific virion morphotypes (Prangishvili et al., 2017).
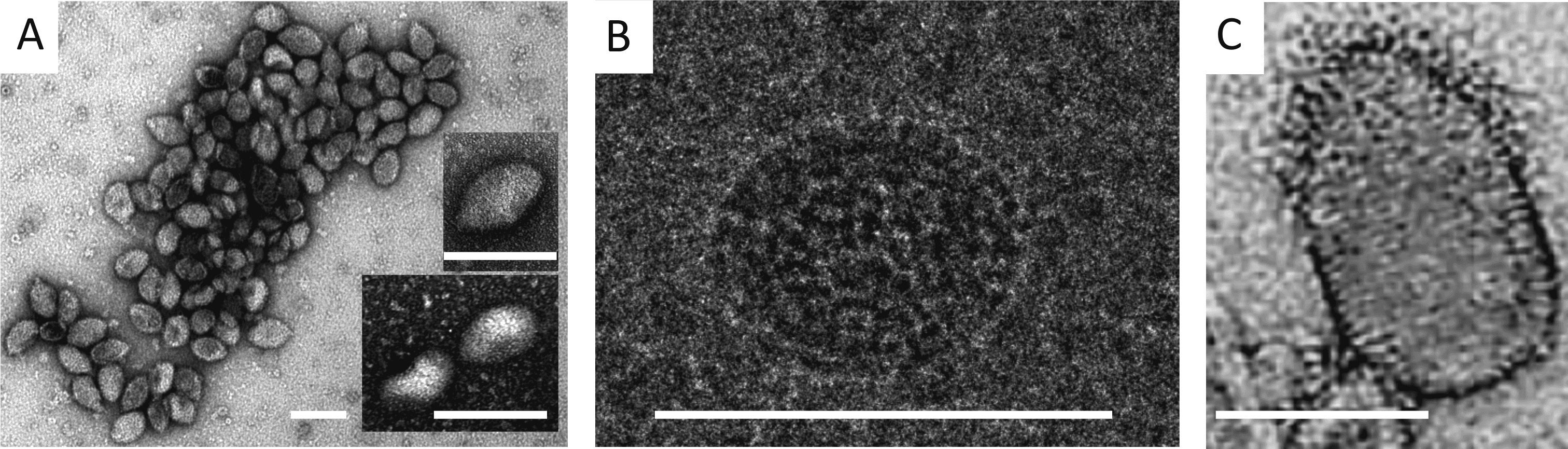 |
| Figure 1 Guttaviridae. Virions of Aeropyrum pernix ovoid virus 1 (A and B) and Sulfolobus newzealandicus droplet-shaped virus (C). Virions were negatively stained (A, C) and embedded in ice (B). Scale bars, 100 nm. (A and B, reproduced with permission from (Mochizuki et al., 2011); (C, Courtesy of W. Zillig) |
Physicochemical and physical properties
Virion densities have not been determined because virions are unstable in CsCl and lyse.
Nucleic acid
Covalently closed circular dsDNA of 13 769 bp.
Proteins
APOV1 virions include one major capsid protein of about 10.5 kDa and at least 2 minor capsid proteins (Mochizuki 2012).
Lipids
Guttaviruses appear to be enveloped but the exact lipid composition is not known.
Carbohydrates
Not known.
Genome organization and replication
The dsDNA genome of APOV1 is 13 769 bp (56.5% GC) and contains 21 ORFs that could encode proteins of more than 56 amino acids, including an integrase of the tyrosine recombinase superfamily, a DnaA-like ATPase, a glycosidase and several DNA-binding proteins containing the helix-turn-helix motifs (Figure 2 Guttaviridae). In most cases, the ORFs are preceded by recognizable putative ribosome binding sites. The majority of putative genes (14) are located on one strand of the DNA. Of the seven putative genes on the opposite strand, five are clustered together (Figure 2 Guttaviridae). Two of the putative genes show sequence similarity to genes of members of the family Fuselloviridae encoding an integrase and a DnaA-like protein, whereas three other genes, including the one for glycosidase, are most closely related to those encoded by hyperthermophilic crenarchaea. APOV1 does not carry a gene for a DNA polymerase, suggesting that its genome is replicated by the host replisome. The DnaA-like ATPase of APOV1 is distantly related to the bacterial DNA replication initiator DnaA and, thus, might be involved in the initiation of the APOV1 genome replication. However, experimental evidence to support this prediction is missing.
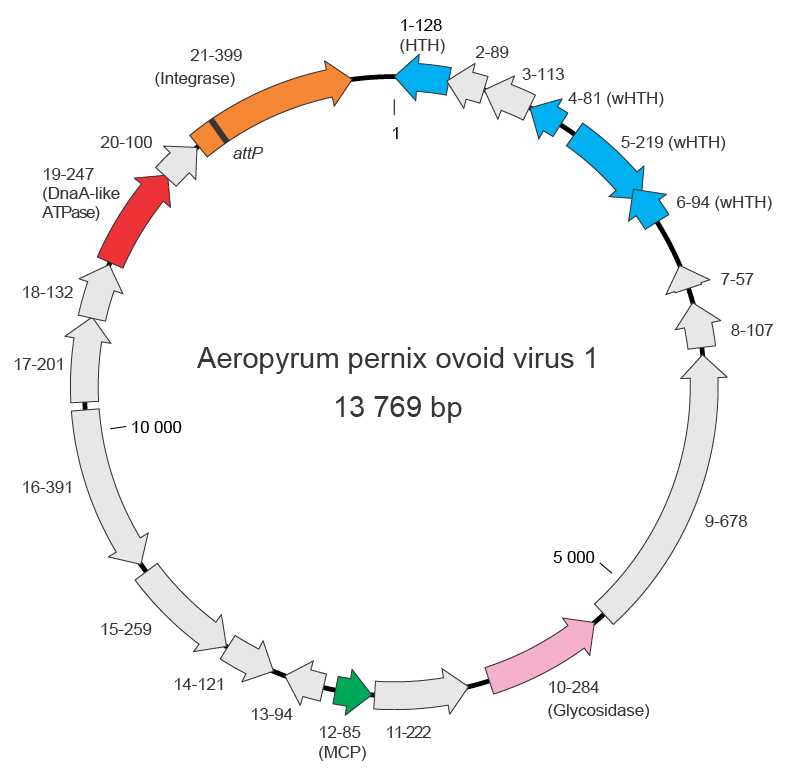 |
| Figure 2 Guttaviridae. Genome map of Aeropyrum pernix ovoid virus 1. Functionally annotated open reading frames are highlighted with different colours. The names of the genes include the information on the length and function (when available) of the encoded proteins. Abbreviations: (w)HTH, (winged) helix-turn-helix DNA-binding proteins; MCP, major capsid protein. |
Biology
Aeropyrum pernix ovoid virus 1 was isolated from Aeropyrum pernix strain K1, which was discovered in a coastal solfataric vent at Kodakara-Jima Island, Japan. The virus is temperate and lysogenizes its host by integrating into the chromosome by recombining with the 3ʹ-distal region of the tRNALeu gene (Mochizuki et al., 2011). The attachment site on the viral genome (attP) is located within the integrase gene. Consequently, upon integration of the APOV1 genome into the host chromosome, the integrase gene is partitioned, as has been also described for fuselloviruses. Excision of the proviral APOV1 genome from the host chromosome, followed by genome replication and virion production, is induced under suboptimal growth conditions, namely, reduced aeration (A.pernix is an obligate aerobe).
Derivation of names
Betaguttavirus: originally the second of two genera in the family, but now the only genus
Guttaviridae: from the Latin gutta, meaning "drop".
Genus demarcation criteria
Given that there is only one genus in the family, genus demarcation criteria have not been set.
Relationships within the family
As only the APOV1 genome sequence is available, there is no analysis of phylogenetic relationships in the family.
Relationships with other taxa
The bipartite gene-sharing network analysis of the archaeal virosphere showed that APOV1 forms a common module with fuselloviruses, indicating that the two groups of hyperthermophilic archaeal viruses might be evolutionarily related (Krupovic et al., 2018, Iranzo et al., 2016). Fuselloviruses and APOV1 share genes for an integrase of the tyrosine recombinase superfamily and a DnaA-like ATPase.
Sulfolobus newzealandicus droplet-shaped virus (SNDV) was the first member of the Guttaviridae, initially classified as the sole representative of the genus Alphaguttavirus. SNDV virions are droplet-shaped, 80 × 130 nm, decorated with dense filaments attached to the pointed end of the virion, and superficially resemble virions of APOV1 (Figure 1C Guttaviridae). SNDV virions have one major capsid protein of about 17.5 kDa (Arnold et al., 2000). Similar to APOV1, SNDV resides within the host cell in a carrier state as an episomal provirus, which is spontaneously induced at the early stationary growth phase. SNDV virion release is associated with host cell lysis (Arnold et al., 2000). The genome is circular dsDNA molecule of about 20 kbp (Arnold et al., 2000). However, the genome of SNDV was not sequenced and the virus has been lost from the collection. Hence, the genus Alphaguttavirus was abolished in 2022.

