Viroids
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Definition
Viroids are small, circular, single stranded, non-protein-coding RNAs that replicate autonomously when inoculated into higher plants. Some are pathogenic, but others replicate without eliciting symptoms in susceptible plant species. Unlike the genomes of conventional viruses, viroids are not protected by a protein capsid.
Physicochemical and physical properties
Viroid molecules, with a Mr of 80–125×103, display extensive internal base pairing and assume, in most cases, a rod-like or quasi-rod-like conformation in vitro that is about 50 nm in length (Figure 1). For some viroids, there is indirect evidence supporting this type of structure in vivo.
These structures denature by cooperative melting (Tm in 10 mM Na+ at about 50 °C) to single stranded circles of about 100 nm contour length. Viroids can also form metastable structures with hairpins that may be functionally important. However, at least two viroids, peach latent mosaic viroid (PLMVd) and chrysanthemum chlorotic mottle viroid (CChMVd), do not follow this rule, and their RNAs clearly appear to adopt branched conformations. There is experimental support for the contention that PLMVd and CChMVd have unique conformations because unlike other viroids, they are insoluble in 2 M LiCl. Some viroids also contain elements of tertiary structure like kissing-loops. Sequences vary from 246 to 401 nt in length and are rich in G+C (53–60%), except avocado sunblotch viroid (ASBVd) which has 38% G+C.
Genome organization and replication
All viroids except ASBVd, PLMVd, CChMVd and eggplant latent viroid (ELVd) share a model of five structural functional domains within the proposed rod-like secondary structure of minimal free energy: C (central), P (pathogenic), V (variable) and TR and TL (terminal right and left). The C domain contains a central conserved region (CCR) formed by two sets of conserved nucleotides located in the upper and lower strands, with those of the upper strand being flanked by an inverted repeat (Figure 2).
The upper strand of the CCR can form either a hairpin or, in oligomers, a double stranded structure possibly relevant in replication. Two other conserved sequence motifs are the terminal conserved region (TCR), found in all members of the genera Pospiviroid and Apscaviroid and in the two largest members of the genus Coleviroid, and a terminal conserved hairpin (TCH) present in all members of the genera Hostuviroid and Cocadviroid (Figure 2). The conservation of these two motifs in sequence and location in the left terminal domain suggests their involvement in some critical functions. Coconut cadang-cadang viroid (CCCVd) is unusual in occurring as RNAs of different sizes, the larger ones having sequence repetitions of the smallest one; a similar situation has been found in citrus exocortis viroid (CEVd). Some viroids, the most striking examples being columnea latent viroid (CLVd) and australian grapevine viroid (AGVd), appear to have arisen by intermolecular RNA recombination events since they seem to consist of a more or less complex mosaic of sequences present in other viroids. Direct evidence in support of an ongoing recombination process has come from studies on the dynamic distribution over time of Coleus viroids coinfecting individual plants.
There is no evidence that viroids encode proteins and unlike viruses that primarily parasitize the host translation machinery, viroids mainly parasitize host transcription, possibly by subverting either nuclear RNA polymerase II (family Pospiviroidae), or one of the chloroplastic RNA polymerases (family Avsunviroidae) to accept RNA templates. Oligomers isolated from infected tissue may be replicative intermediates produced by a rolling circle mechanism with two variants (asymmetric and symmetric) and three steps (RNA polymerization, cleavage and ligation). Initiation of RNA synthesis in PSTVd and ASBVd appears to occur at specific sites, suggesting that it is promoter-driven. In the family Avsunviroidae oligomeric RNAs self-cleave in vitro and very probably in vivo through hammerhead structures to produce unit-length strands, but in the family Pospiviroidae cleavage is catalyzed by host factors (probably one or more of the RNase III-like proteins) (Figure 3). Ligation of PSTVd appears to be mediated by a nuclear RNA ligase, whereas this reaction has been proposed to be autocatalytic for PLMVd leading to unusual 2′, 5′-phosphodiester bonds at the ligation sites; alternatively, a chloroplastic RNA ligase might mediate this step.
Antigenic properties
No antigenicity demonstrated.
Biological properties
Host range
Some viroids have wide host ranges in the angiosperms but others, particularly members of the family Avsunviroidae, have narrow host ranges. CCCVd and coconut tinangaja viroid (CTiVd) infect monocotyledons. Old cultivars of grapevine and citrus can harbor at least five different viroids. A single nucleotide substitution converts PSTVd from a non-infectious RNA to one that is infectious for Nicotiana tabacum.
Symptoms
Some viroids have devastating effects, for example, CCCVd has killed millions of coconut palms in the Philippines, while others cause epinasty, rugosity, chlorosis and necrosis on leaves, internode shortening of stems leading to stunted plants, bark cracking, deformation and color alterations of fruits and storage organs, and delays in foliation, flowering and ripening. A few viroids induce only mild or no symptoms. Symptom expression is generally more pronounced at high temperature and light intensities.
Molecular determinants of specific symptoms have been mapped in the genome of members of both families. Small sequence changes can convert a severe strain into a symptomless strain (or vice versa). Direct interaction of the genomic viroid RNA with host factors, and involvement of or interference with the plant RNA silencing machinery, have been suggested as primary event(s) of symptom induction.
Transmission
Viroids of horticultural species are transmitted mainly by vegetative propagation. In plants propagated via seeds, they may be transmitted mechanically or through seed or pollen. Only tomato planta macho viroid (TPMVd) is known to be efficiently transmitted by aphids.
Movement
To invade plants systematically, viroids must be able to move from the initially infected cells to the surrounding ones and then to the vascular system. Three different types of movement can be considered: intracellular, cell-to-cell and long-distance. PSTVd movement into the nucleus appears to be a cytoskeleton-independent process that is mediated by a specific and saturable receptor and involves recognition of a conserved sequence and/or structural motif in the upper central conserved region. How members of the family Avsunviroidae are transported into chloroplasts is unknown. Mutational analysis of PSTVd has shown that many of the loops in its rod-like secondary structure (see Figure 6) play a role in movement from cell-to-cell and across tissue boundaries, possibly by creating pockets that bind host proteins. In vitro, hop stunt viroid (HpSVd) can form a ribonucleoprotein complex with the phloem protein 2, a dimeric lectin able to move from cell-to-cell through plasmodesmata and toward sink tissues in the assimilate stream. These properties, together with its ability to bind RNA, suggest that this lectin facilitates the systemic movement of viroids. Access of PSTVd to floral and vegetative meristems is impaired, most likely by an RNA silencing mechanism. In contrast, a chloroplast replicating viroid, PLMVd, is able to enter the apical meristem of its natural host.
Cross protection
Interactions at the level of symptom expression and viroid accumulation have been detected in plants co-infected by two strains of a viroid or even by two different viroids sharing extensive sequence similarities. Interactions of this class have been observed in the case of members belonging to both viroid families (see below), suggesting the possible existence of more than one mechanism of cross-protection between viroids. Alternatively, because RNA-mediated cross-protection in plant viruses is mechanistically similar to post-transcriptional gene silencing, cross-protection between viroids might occur through a similar mechanism in both families.
Phylogenetic relationships within the viroids
Criteria such as the nuclear site of replication and accumulation, the presence and type of CCR, and the presence or absence of the two other conserved regions TCR and TCH, are used for classifying most viroids in family Pospiviroidae, with the two latter criteria serving for their allocation into genera. ASBVd, PLMVd, CChMVd and ELVd form a second family of viroids, the Avsunviroidae, without CCR but endowed with the ability to self-cleave via hammerhead ribozymes (Figure 3). The type of hammerhead structure, together with the G+C content and the solubility in 2 M LiCl are used to demarcate species into genera. Essentially the same groupings are obtained by using phylogenetic trees derived from the whole sequences (Figure 4).
Species demarcation criteria
An arbitrary level of less than 90% sequence similarity and distinct biological properties, particularly host range and symptoms, currently separate most species within each viroid genus. This second criterion is now mandatory for the creation of new viroid species. Where host range is restricted to a single botanical species not expressing symptoms, other distinct biological properties should be described (i.e. seed transmission, movement and distribution within the host, differential fitness in competition assays) to justify creation of a new viroid species. Due to the heterogeneous nature of viroid populations, infected plants often contain a spectrum of closely related variants, each showing more than 90% sequence similarity. One or a limited number of these variants may represent the bulk of the population.
List of unassigned viroids which have not been approved as species
| Blueberry mosaic viroid-like RNA | (BluMVd-RNA) |
| Burdock stunt viroid | (BuSVd) |
| Nicotiana glutinosa stunt viroid | (NGSVd) |
| Pigeon pea mosaic mottle viroid | (PPMMoVd) |
| Tomato bunchy top viroid | (ToBTVd) |
Derivation of names
Viroid: from the name given to the subviral agent of potato spindle tuber disease.
Apsca: from apple scar skin viroid.
Avsun: avocado sunblotch viroid.
Cocad: from coconut cadang-cadang viroid.
Cole: from Coleus blumei viroid 1.
Ela: from eggplant latent viroid.
Hostu: from hop stunt viroid.
Pelamo: from peach latent mosaic viroid.
Pospi: from potato spindle tuber viroid.
Further reading
Journals and books
[1] F. Bussière, J. Ouellet, F. Coté, D. Lévesque, J.P. Perreault., Mapping in solution shows the peach latent mosaic viroid to possess a new pseudoknot in a complex, branched secondary structure. J. Virol. 74 (2000) 2647–2654.
Diener, 1971 T.O. Diener, Potato spindle tuber “virus” IV. A replicating low molecular weight RNA. Virology. 45 (1971) 411–428.
Ding, 2009 B. Ding, The biology of viroid–host interactions. Annu. Rev. Phytopathol. 47 (2009) 105–131.
Elena et al., 2001 S.F. Elena, J. Dopazo, M. De la Peña, R. Flores, T.O. Diener, A. Moya, Phylogenetic analysis of viroid and viroid-like satellite RNAs from plants: a reassessment. J. Mol. Evol. 53 (2001) 155–159.
Flores et al., 2005 R. Flores, C. Hernández, A.E. Martínez de Alba, J.A. Daròs, F. Di Serio, Viroids and viroid–host interactions. Annu. Rev. Phytopathol. 43 (2005) 117–139.
Gas et al., 2007 M.E. Gas, C. Hernández, R. Flores, J.A. Daròs, Processing of nuclear viroids in vivo: An interplay between RNA conformations. PLoS Pathog. 3 (2007) 1813–1826.
Gross et al., 1978 H.J. Gross, H. Domdey, C. Lossow, P. Jank, M. Raba, H. Alberty, H.L. Sänger, Nucleotide sequence and secondary stucture of potato spindle tuber viroid. Nature. 273 (1978) 203–208.
Hadidi et al., 2003 A. Hadidi, R. Flores, J.W. Randles, J.S. Semancik, Viroids. In: A. Hadidi, R. Flores, J.W. Randles, J.S. Semancik, Viroids. CSIRO Publishing, Collingwood, Australia2003.
Keese and Symons, 1985 P. Keese, R.H. Symons, Domains in viroids: evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl Acad. Sci., U S A. 82 (1985) 4582–4586.
Owens, 2007 R.A. Owens, Potato spindle tuber viroid: The simplicity paradox resolved?. Mol. Plant Pathol. 8 (2007) 549–560.
Zhong et al., 2008 X. Zhong, A.J. Archual, A.A. Amin, B. Ding, A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell. 20 (2008) 35–47.
Websites
Subviral RNA Database: http://subviral.med.uottawa.ca.
NCBI Entrez Viral Genomes (includes viroids): http://www.ncbi.nlm.nih.gov/genomes/GenomesHome.cgi?taxid=10239.
Contributed by
Owens, R.A., Flores, R., Di Serio, F., Li, S-F., Pallás, V., Randles, J.W., Sano, T. and Vidalakis, G.
Figures
Figure 1 Electron micrograph of potato spindle tuber viroid (PSTVd) and viral DNA (coliphage T7 DNA) illustrating the comparative sizes and the rod-like structure of the viroid. The scale bar represents 0.5 m.
(From Diener, T.O. (1979). Viroids and Viroid Diseases. Wiley, New York; with permission.)
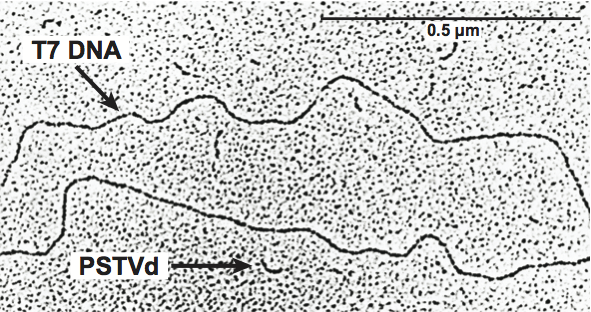
Figure 2 Rod-like structure models for viroids in the different genera of the family Pospiviroidae. The type species of each genus is indicated on the right and the approximate location of the five domains C (central), P (pathogenic), V (variable) and TL and TR (terminal left and right), at the top of the figure. The core nt of the central conserved region (CCR), terminal conserved region (TCR) and terminal conserved hairpin (TCH), are shown. Arrows indicate flanking sequences which form, together with the core nucleotides of the CCR upper strand, imperfect inverted repeats. The core nucleotides of the hop stunt viroid (HpSVd) CCR have been defined by comparison with columnea latent viroid (CLVd), which on the basis of other criteria (presence of the TCR, absence of the TCH and overall sequence similarity) is included in the genus Pospiviroid. Substitutions found in the CCR and TCR of iresine viroid 1 (IrVd-1) a member of the genus Pospiviroid, are indicated in lowercase. In the genus Coleviroid the TCR only exists in the two largest members coleus blumei viroid 2 (CbVd-2) and coleus blumei viroid 3 (CbVd-3).
(Adapted from Flores, R. et al. (1997). Viroids: the non-coding genomes. Semin. Virol., 8, 6573.)
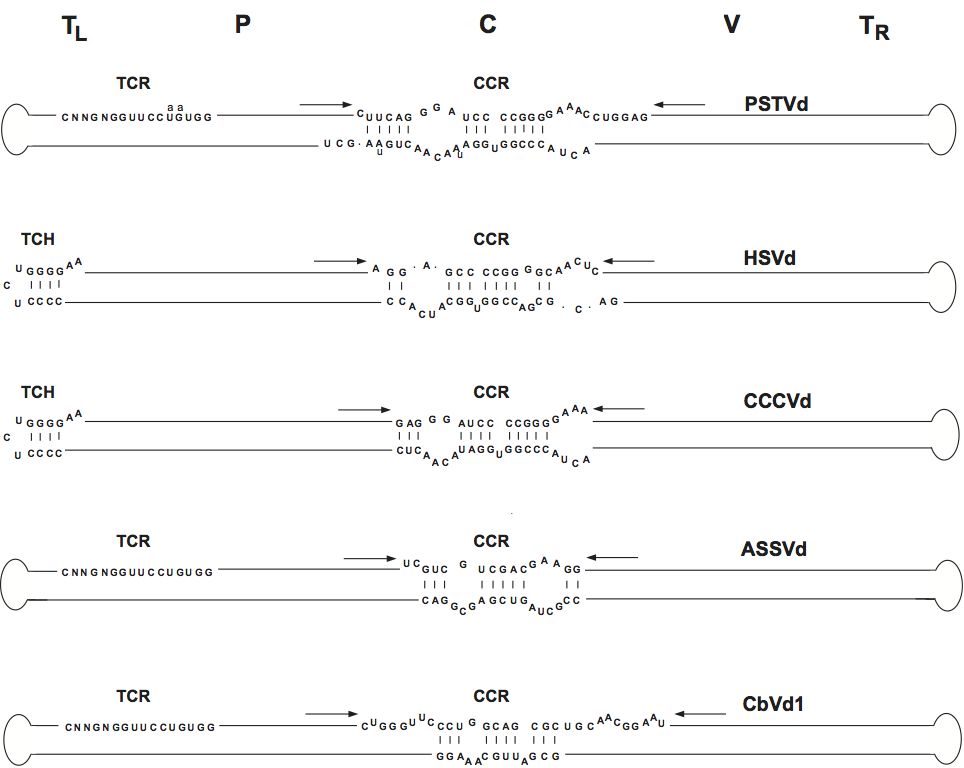
Figure 3 Models for viroid replication. The plus polarity (solid lines) is assigned by convention to the most abundant infectious RNA and the minus polarity (open lines) to its complementary strand. The alternative asymmetric and symmetric pathways involve one and two rolling circles, respectively. In the symmetric variant, cleavage of plus and minus multimeric strands is mediated by hammerhead ribozymes (RZ), which lead to linear monomeric RNAs with 5-hydroxyl and 2-3-cyclic phosphodiester termini. Arrowheads denote the cleavage sites. The hammerhead structures that can be formed by avocado sunblotch viroid (ASBVd), peach latent mosaic viroid (PLMVd) and chrysanthemum chlorotic mottle viroid (CChMVd) RNAs are shown on the right; conserved nucleotides are boxed. In the asymmetric variant, cleavage of plus multimeric strands relies on a host factor (HF), which generates linear monomeric RNA containing probably 5-phosphomonoester and 2 and 3-hydroxyl termini.
(Adapted from Flores, R. et al. (1997). Viroids: the non-coding genomes. Semin. Virol., 8, 6573.)
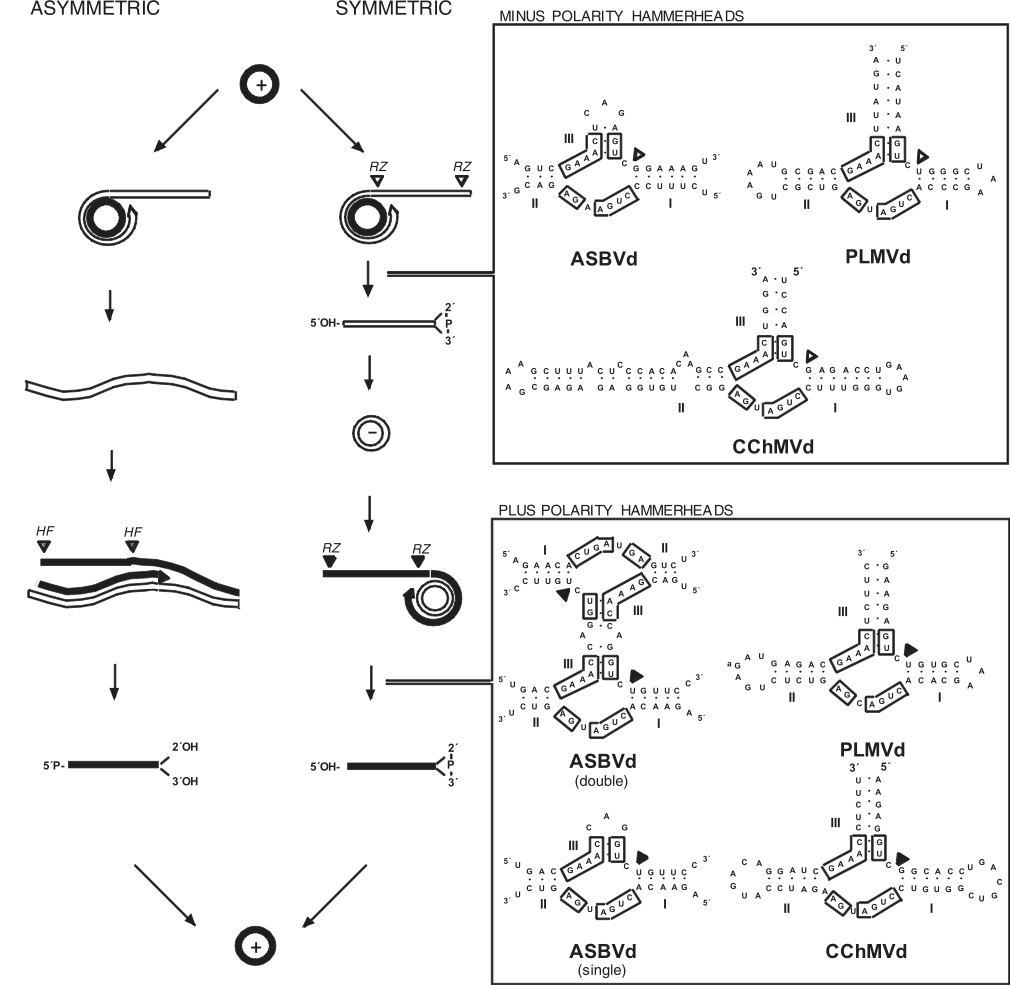
Figure 4 Consensus phylogenetic tree (based on 1000 replicates) obtained for viroids. Isolates indicated by ***, ** and * form monophyletic groups in more than 95%, 85% and 75% of the replicates, respectively. Numbering of sequences was based on the similarities found between the upper strands of the central conserved regions of members of the family Pospiviroidae and the upper-left-hand portions of the hammerhead structures of members of the family Avsunviroidae (Diener, T.O. (1991) Subviral pathogens of plants: viroids and viroidlike satellite RNAs. FASEB J., 5, 28082813). The alignment was performed with the Clustal X program, version 1.64 (gap opening and gap extension penalties 15 and 1, respectively) and minor manual adjustments between the conserved regions. Evolutionary distances were estimated according to the model of Jukes and Cantor and the phylogenetic tree was constructed by the neighbor-joining method using the MEGA program (version 1.01). (De la Pea and Flores, unpublished data.)
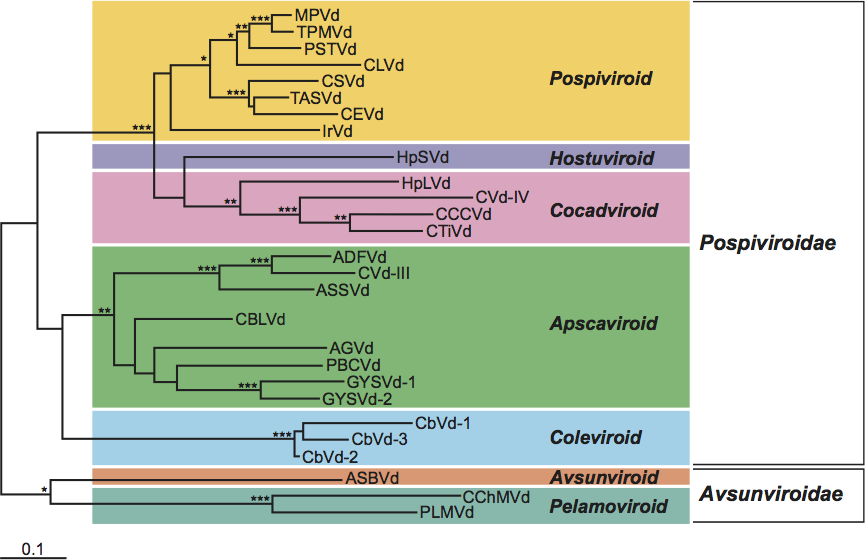
Figure 5 Primary and proposed branched secondary structure of lowest free energy for the reference sequence variant of an isolate of peach latent mosaic viroid. Plus and minus self-cleavage domains are delimited by flags, the 13 conserved residues present in most of the hammerhead structures are indicated by bars, and the self-cleavage sites are shown by arrows. Solid and open symbols refer to plus and minus polarities respectively. In vitro chemical and enzymatic probing has revealed the probable existence of a kissing-loop interaction between residues GCGG and CCGC (position 178181 and 211214, respectively) (Bussire et al., 2000).
(Adapted from Hernndez, C. and Flores, R. (1992). Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl Acad. Sci., U S A, 89, 37113715.)
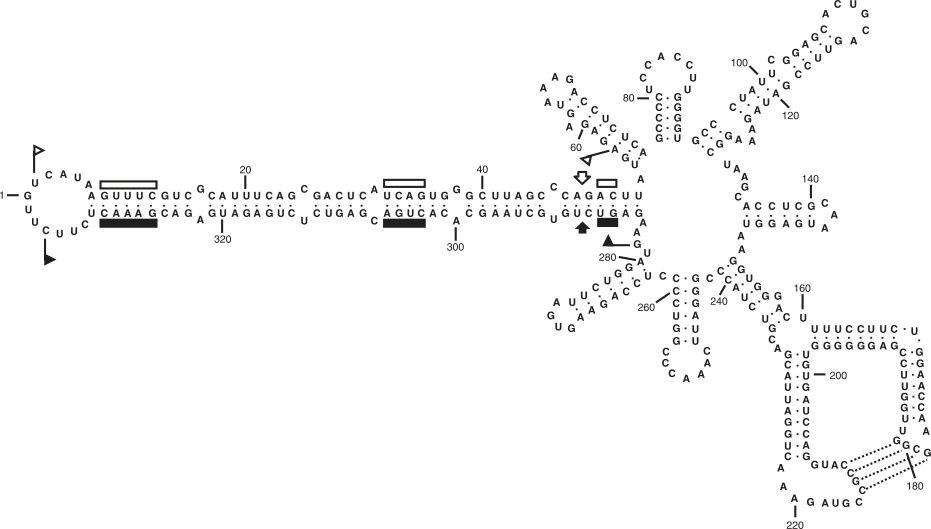
Figure 6 Primary and proposed rod-like secondary structure of lowest free energy for the reference sequence variant of potato spindle tuber viroid (PSTVd).
(Adapted with modifications from Gross, H.J. et al. (1978). Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature, 273, 203208.)

