Fungal Prions
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Taxonomic structure of the fungal prions
Prions
[URE3] prion
[PSI] prion
[Het-s] prion
[PIN] prion
[β] prion
[SWI] prion
[OCT] prion
[MOT3] prion
[ISP] prion
Distinguishing features
The name “prion” means infectious protein. In yeast and filamentous fungi, infection by viruses occurs exclusively by cell–cell fusion with transmission of cytoplasm. Viruses are transmitted as non-chromosomal genetic elements (denoted in brackets as above). Likewise, one expects prions of these organisms to show similar behavior. There are now eight well-documented prions in yeast and filamentous fungi. Seven of these are due to self-propagating amyloids, while the eighth is an auto-activating protease.
Prion properties
Morphology
Prions are infectious proteins: altered forms of a normal cellular protein that may have lost their normal function, but have acquired the ability to change the normal form of the protein into the same abnormal form as themselves. In most cases they form filaments or filamentous aggregates, but this is not part of the definition of a prion, which in some cases could be based on a self-perpetuating covalent modification without substantial morphological effects. Amyloid filaments (Figure 1) are typically straight and unbranched. Some have a helical appearance. In several cases, amyloid formed in vitro of the recombinant prion protein has been shown to be infectious for yeast, transmitting the corresponding prion.
Physicochemical and physical properties
Except for [β], each prion is a self-propagating amyloid form of the corresponding protein (see below). Infectious amyloids of the prion domains of Ure2p, Sup35p and Rnq1p have an in-register parallel β-sheet architecture (Figure 2). Infectious amyloid of the HET-s prion domain is a two-turn β-helix. The vacuolar protease B is processed into its mature active form in cells carrying the [β] prion; the [β] prion is simply the mature active form of protease B.
Nucleic acid
None. The definition of a prion is that it is an infectious protein without the need for any accompanying nucleic acid, although the host genome must encode the protein.
Proteins
| Prion | Protein | Function of the protein |
| [URE3] | Ure2p | Regulator of nitrogen catabolism |
| [PSI] | Sup35p | Translation termination |
| [Het-s] | HET-s | Heterokaryon incompatibility |
| [PIN] | Rnq1 | Unknown; detected by [PSI]-inducibility |
| [β] | Prb1p | Protease B: protein processing and degradation |
| [SWI] | Swi1p | Subunit of chromatin remodeling complex |
| [OCT] | Cyc8p | Transcription repressor with Tup1p |
| [MOT3] | Mot3p | Transcription factor |
| [ISP] | Sfp1 | Transcription factor |
Lipids
None.
Carbohydrates
None.
Genome organization and replication
Each of the prions is inherited as a non-Mendelian genetic element (also called cytoplasmic genetic elements or non-chromosomal genetic elements). The “genetic material” in this case is the altered protein which is self-replicating by changing the normal form of the protein (which nonetheless has the same primary sequence) into the altered form. Thus altered Ure2p, Sup35p, Rnq1p, Prb1p, HETs, Swi1p, Cyc8p, Mot3p and Sfp1p are the “genomes”of [URE3], [PSI], [PIN], [β], [Het-s], [SWI], [OCT], [MOT3] and [ISP].
Each amyloid-forming prion protein has a prion domain. Deletions of the remainder of the molecule increase the frequency with which the prion form arises. Chaperones play an important role in all amyloid-related prions of S. cerevisiae, but different prions show different effects of altered chaperone activities. The propagation of all amyloid-based yeast prions requires Hsp104, and its cooperating chaperones, for the breakage of large fibers into smaller ones, to create new “seeds” (Figure 3).
Biological properties
The yeast and fungal prions are passed from cell to cell during mating and hyphal anastomosis. There are no known natural vectors or extracellular transmission. Thus, fungal prions appear first as non-Mendelian genetic elements. Three biologic (genetic) criteria have been proposed to determine whether a given non-Mendelian genetic element is a prion (Figure 4).
- Prions are “reversibly curable”, meaning that even if a prion can be cured from a strain, it can arise again spontaneously at some low frequency in the cured strain.
- Overexpression of the normal protein increases the frequency with which the prion form arises.
- The phenotype of the presence of the prion is the same as the phenotype of a recessive mutant in the gene encoding the protein. This gene first appears as a chromosomal gene necessary for the propagation of the prion. In some cases, the prion produces a phenotype by having an action beyond inactivation of the normal form. In this case, the phenotypic relation does not provide evidence that the non-Mendelian genetic element is a prion, but, of course, it is also not evidence against it being a prion. In all cases, the prion depends for its propagation on the gene for the protein.
Further, physical or chemical evidence for an alteration of the protein in cells carrying the prion should be demonstrated.
The [Het-s] and [β] prions are advantageous for their hosts and are present in essentially all of the respective wild-type strains. Most of the other prions produce a phenotype resulting from a deficiency of the normal protein. However, the [PIN] prion phenotype of priming generation of the [PSI] or [URE3] prion is not produced by deficiency of the corresponding protein, Rnq1p.
A feature of most prions is the existence of several prion “variants” (called “prion strains” in mammals) with different biological properties although the sequence of the prion protein is identical. The differences include phenotype intensity, stability of the prion, sensitivity to overproduction or deficiency of certain chaperones, and height of species barriers.
Species demarcation criteria
Prions are divided into species based on the identity of the protein that makes up the infectious element. (Note: The formal taxonomy of viruses does not extend to unconventional agents like prions and prion “species” are not included in the ICTV Master Species List.)
Species [URE3]
Host Saccharomyces cerevisiae
Distinguishing features
[URE3] is a self-propagating amyloid form of the Ure2 protein, a regulator of nitrogen catabolism. In the prion form of the Ure2 protein, its nitrogen regulation activity is largely lost. This results in a slow growth phenotype as well as derepression of activities normally repressed by a good nitrogen source.
Prion properties
Morphology
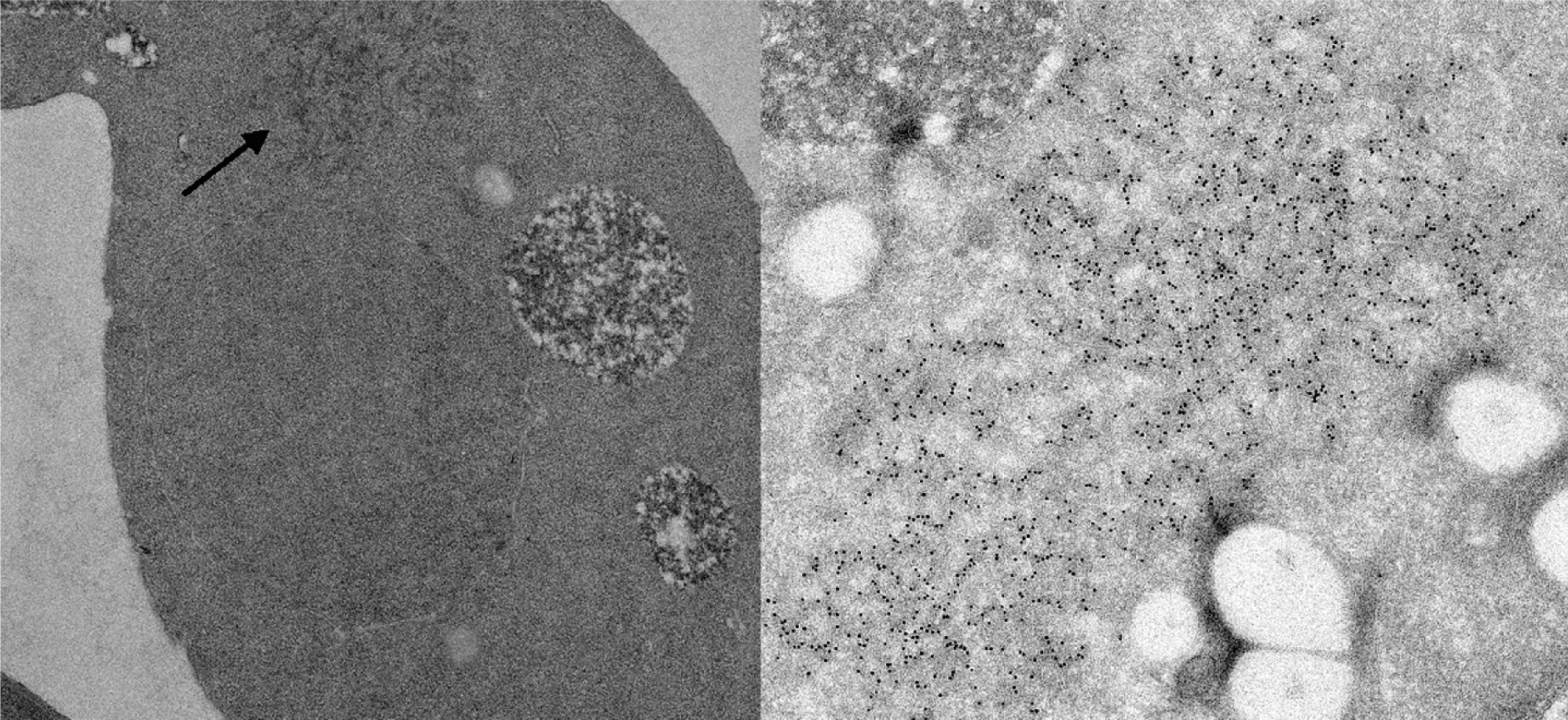
The synthetic prion domain of Ure2p, Ure2p1-65, forms amyloid in vitro, and induces a self-propagating amyloid formation by full-length native Ure2p purified from yeast. Ure2p amyloid filaments have a 4 nm core composed of the prion domain, surrounded by globular appendages composed of the C-terminal nitrogen regulation domain. Cells carrying the [URE3] prion contain a network of 25 nm diameter filaments containing the Ure2 protein (Figure 5).
Physicochemical and physical properties
Ure2p is normally a soluble dimer, whose C-terminal domain is similar in sequence and structure to glutathione - S - transferases. Ure2p is more proteinase K-resistant in extracts of [URE3] strains than in wild-type strains, with the N-terminal prion domain the most protease resistant part. Ure2p amyloid formed in vitro likewise has a core composed of the prion domain which is the most protease-resistant part of the aggregate. The amyloid core of the filaments has beta-sheet structure. Solid-state NMR of prion domain amyloid filaments shows that they have an in-register parallel β-sheet structure. The prion domain changes from being unstructured in the normal soluble form to being in a very tight in-register parallel β-sheet structure in the amyloid (prion) form. The C-terminal domain does not change significantly in structure on conversion from soluble to amyloid form.
Proteins
Ure2p, in its altered form, is the primary component of [URE3]. The presence of the [PIN] prion also stimulates the rate of [URE3] prion generation.
Genome organization and replication
The URE2 gene has an ORF coding for a 354 aa polypeptide, of which the C-terminal part, including residues 81 to 354, is capable of carrying out the nitrogen regulation function of Ure2p if overexpressed. The N-terminal 80 residues (the prion domain) is necessary for in vivo stability of Ure2p and for tight nitrogen regulation. The Ure2p prion domain can propagate [URE3] in the absence of the C-terminal domain and efficiently transmits the prion to the full-length molecule. Amyloid of the prion domain alone or fused to another unrelated protein is highly infectious, transmitting the [URE3] to yeast cells. The prion domain is required for induction of [URE3], for propagation of the prion, and without a covalently attached prion domain, a Ure2 molecule is not affected by the presence of [URE3]. Propagation of [URE3] requires the chaperones Hsp104 and Ssa2 and is blocked by overproduction of the Hsp40-group chaperone Ydj1p, and by the GTP-exchange protein Sse1p. Overexpression of Btn2p also cures [URE3], apparently by collecting [URE3] seeds to one cellular site. Deletion of Btn2 increases the number of [URE3] seeds per cell.
Antigenic properties
Not studied.
Biological properties
[URE3] makes cells able to take up ureidosuccinate from the media containing a good nitrogen source such as ammonia or glutamine. The ure2 mutants have the same phenotype, and the propagation of [URE3] requires the URE2 gene. Yeast cells turn off transcription of genes for enzymes involved in utilization of poor nitrogen sources when a good nitrogen source is available (nitrogen catabolite regulation). DAL5 encodes the permease for allantoate, a poor, but usable, nitrogen source for yeast. DAL5 is thus subject to nitrogen catabolite regulation. Allantoate is structurally similar to ureidosuccinate, and so Dal5p can take up ureidosuccinate. Ureidosuccinate is an intermediate in uracil biosynthesis, the product of the first step in the pathway, aspartate transcarbamylase (URA2). Thus, ura2 mutants can grow on ureidosuccinate in place of uracil if the cell has either the [URE3] prion or a mutation in ure2. [URE3] can be cured by growth in the presence of 5 mM guanidine HCl.
[URE3] is not found in wild strains indicating that [URE3] is a disease of yeast rather than an adaptive mechanism. Indeed, one Saccharomyces species has an N-terminal domain similar to the cerevisiae Ure2 prion domain, but is unable to have the [URE3] prion, indicating that this domain is not present in order to allow cells to have a prion. The prion domain has a normal function in stabilizing the full-length protein against degradation, again consistent with [URE3] being a disease. The prion domain changes in evolution more rapidly than the C-terminal part of the molecule resulting in a species barrier to [URE3] transmission. This variation may be selected to protect cells from acquiring the prion. Finally, [URE3] cells generally grow slowly – not an advantageous trait.
Prion variants
Even a single protein can form prions of distinct “strains”, or “variants”, apparently due to different self-propagating amyloid structures, much as a given protein can assume more than one crystal form. Variants are distinguished by the intensity of their phenotypes, by the stability of prion propagation, by the effect of overproduction or deficiency of various chaperones on prion propagation and by species barriers. Different yeast species can have the [URE3] prion, but their differing Ure2p sequences produce a “species barrier” to transmission. Saccharomyces castellii is apparently unable to have the [URE3] prion, while S. bayanus, S. cariocanus and S. mikatae can have it.
Species [PSI+]
Host Saccharomyces cerevisiae
Distinguishing features
[PSI] is a self-propagating amyloid form (a prion) of the Sup35 protein, a subunit of the translation termination factor of the yeast Saccharomyces cerevisiae. The presence of the [PSI] prion results in a relative shortage of the soluble form of Sup35p, and a resulting decrease in the efficiency of translation termination. This makes readthrough of termination codons more frequent. The phenotype of [PSI] is ability to grow in the absence of adenine in spite of a premature stop mutation in ADE1 or ADE2, combined in the latter case with a weak serine-inserting tRNA suppressor mutation. The prion domain of Sup35p (residues 1-123) also is necessary for normal mRNA turnover, interacting with the polyA binding protein and the polyA-degrading enzymes to favor shortening of the 3′ polyA structure of mRNAs, a first step in mRNA degradation.
Prion properties
Morphology
Sup35p forms typical amyloid filaments in vitro (Figure 1), and well-ordered, laterally aligned arrays of filaments in vivo.
Physicochemical and physical properties
Amyloid filaments of the Sup35 protein are high in beta sheet structure, but are only slightly protease-resistant. They have an in-register parallel β-sheet structure based on solid-state NMR studies. The filaments have one molecule for about every 4.7 Å of filament length, consistent with the in-register parallel structure. Filaments formed under different conditions may have different extents of β-sheet structure in the prion domain.
Proteins
Sup35p (79 kDa) is a subunit of the translation termination factor, which also includes Sup45p. Amyloid of Sup35p isolated from [PSI]-containing cells also contains the Hsp70 group chaperone Ssa1/2 in large amounts and smaller amounts of Hsp104, Ssb1/2, Sis1, Sse1, Ydj1 and Sla2.
Genome organization and replication
The N-terminal 123 residues of Sup35p are sufficient to propagate the [PSI] prion. This part of Sup35p is very high in glutamine and asparagine residues, and these are critical for prion formation and propagation. Overexpression of this prion domain of Sup35p also induces the de novo appearance of [PSI] at a much higher frequency than does similar overexpression of the full-length Sup35p. Amyloid of full-length Sup35p or the prion domain (with or without fusion to another protein) is infectious for yeast transmitting the [PSI+] prion.
The level of the Hsp104 chaperone is critical for [PSI] propagation with either overexpression or under-expression leading to the loss of the prion. Hsp104, together with other chaperones, cleaves long filaments to make shorter ones that can serve as new seeds of amyloid formation. The Hsp70s in the Ssa group are also essential for [PSI] propagation.
Antigenic properties
Not studied.
Biological properties
[PSI] increases the efficiency with which ribosomes read through termination codons. The Sup35 protein is a subunit of the translation release factor that recognizes termination codons, and releases the peptidyl tRNA and cleaves the nascent peptide from the tRNA. Thus, sup35 mutants have the same phenotype as do [PSI] strains. Furthermore, the SUP35 gene is necessary for the propagation of [PSI]. [PSI], like [URE3] and [PIN], can be cured by guanidine inhibition of or deficiency of Hsp104, but from the cured strains, can again be isolated subclones carrying [PSI]. Hsp104 is a disaggregating chaperone necessary for the propagation of each of these prions. By breaking up amyloid filaments into smaller filaments, Hsp104 increases the number of “seeds” and so ensures inheritance of the amyloid by each of the daughter cells. Overproduction of Sup35p results in a 100-fold increase in the frequency with which [PSI] arises de novo.
Strains of [PSI], like the strains known for the scrapie agent and other transmissible spongiform encephalopathies, have been found for [PSI]. They are associated with different stabilities, curability by guanidine, and efficiency of suppression of nonsense mutations.
[PSI+] is not found in wild strains, suggesting that it is a disease of yeast, rather than an adaptive mechanism. The prion domain of Sup35p has a normal non-prion function in mRNA turnover, by facilitating the normal gradual shortening of the 3′ polyA structure. Thus, like the mammalian prion protein PrP, the Sup35 prion domain is not present simply to allow cells to have a prion. The Sup35 prion domain changes more rapidly in evolution than does the C-terminal domain, developing a species barrier to [PSI+] transmission. This variation may be selected to protect against acquisition of a prion. In one study, one-quarter of wild S. cerevisiae isolates had a large deletion in their prion domain so that they could no longer become [PSI+], indicating that prion formation is not conserved within this species.
Prion variants
Even a single protein can form prions of distinct “strains”, or “variants”, apparently due to different self-propagating amyloid structures, much as a given protein can assume more than one crystal form.
Species [PIN+]
Host Saccharomyces cerevisiae
Distinguishing features
[PIN+] is a self-propagating amyloid form of the Rnq1 protein, whose normal function is unknown. [PIN+] has the ability to dramatically increase the frequency with which the [PSI+] prion arises. [PIN+] also has a smaller effect on [URE3] prion generation.
Prion properties
Morphology
Rnq1p is aggregated in [PIN+] strains and recombinant Rnq1p forms amyloid filaments in vitro.
Physicochemical and physical properties
Filaments of the Rnq1 protein formed in vitro are 11 nm in diameter, stain with Thioflavin T, and have high β-sheet content indicating amyloid structure. Solid state NMR studies of the Rnq1p prion domain indicate that the amyloid has an in-register parallel architecture.
Proteins
Rnq1p is a 405 aa residue protein with a C-terminal domain rich in asparagine and glutamine residues (like the prion domains of Ure2p and Sup35p). This domain can act as a prion domain when fused to the C-terminal part of Sup35p. Amyloid of Rnq1p isolated from [PIN+] cells contains roughly equimolar amounts of the Hsp40 family chaperone Sis1p.
Genome organization and replication
Amyloid formed in vitro from recombinant Rnq1p is infectious for yeast transmitting the [PIN+] prion. The Hsp104 chaperone is essential for [PIN] propagation. The Hsp40 Sis1p is also essential for [PIN] propagation.
Antigenic properties
Not studied.
Biological properties
[PIN] dramatically increases the frequency with which the [PSI] prion arises de novo, so much so that it is nearly essential for [PSI] generation. [PIN] also increases the frequency of [URE3] de novo generation, but only 10- to 100-fold. The evidence suggests that the amyloid formed by one asparagine–glutamine-rich protein can prime the formation of amyloid by another such protein. Overproduction of Swi1p or Cyc8p can mimic [PIN+] in stimulating generation of [PSI+], and now these proteins are known to form prions themselves, [SWI+] and [OCT+], respectively.
Prion variants
Even a single protein can form prions of distinct “strains”, or “variants”, apparently due to different self-propagating amyloid structures, much as a given protein can assume more than one crystal form. [PIN+] is known to have several variants distinguished by their strength in inducing [PSI+] formation.
Species [Het-s]
Host Podospora anserina, a filamentous fungus
Distinguishing features
[Het-s] was found as a non-chromosomal gene necessary for heterokaryon incompatibility based on the chromosomal het-s/S locus. [Het-s] is a self-propagating amyloid of the HET-s protein. Heterokaryon incompatibility is a normal fungal function, and so the [Het-s] prion is the first prion known to be responsible for a normal function, a conclusion further confirmed by the fact that nearly all wild het-s strains carry this prion. Amyloid formed in vitro from recombinant HET-s protein is infectious to normal Podospora transmitting the [Het-s] prion.
Prion properties
Morphology
The HET-s protein shows self-seeding formation of amyloid filaments 15–20 nm in diameter.
Physicochemical and physical properties
The amyloid of the HET-s prion domain is a two-turn β-helix with a very uniform structure. This is in contrast to the amyloids of the yeast prions which are heterogeneous in detail structure, although uniform in having in-register parallel architecture. There is only one known [Het-s] prion variant, and this may explain the uniform structure. The HET-s protein has evolved to be a prion with special properties, explaining the uniformity of filament structure.
Proteins
The HET-s protein is the product of the het-s gene. The prion domain sufficient for amyloid formation in vitro and prion propagation in vivo is located at the C-terminal part of the molecule. Deletions of the N-terminal part destabilize the prion domain.
Genome organization and replication
The prion domain of the HET-s protein is the C-terminal 72 aa residues. This region is very protease-sensitive in the soluble form and protease resistant in the amyloid form. Unlike the proteins that form the [URE3], [PSI] and [PIN] prions, the HET-s protein prion domain is not rich in asparagine or glutamine residues. Residues 23 and 33 are particularly critical in determining whether the protein can undergo the prion change or not.
Antigenic properties
Not studied.
Biological properties
[Het-s] makes Podospora anserina strains differing at the het-s locus able to carry out heterokaryon incompatibility. It is the first prion described that is responsible for a normal cellular function, rather than a disease of the organism. When two fungal colonies grow together, the hyphae of the two colonies fuse with each other. This process facilitates cooperation between the colonies in acquisition of nutrients, and perhaps other functions. One risk of this process is that viruses present in one colony will spread into the other colony. Apparently to prevent this, most fungi will only form heterokaryons with closely related strains, identical for alleles at each of 6 or more loci. In Podospora anserina, there are eight such loci called Het loci. The het-s locus has alleles het-s and het-S. The het-s strains show incompatibillity for heterokaryon formation with het-S strains only if the protein encoded by het-s is in a prion form. The presence of this prion is found as a non-Mendelian genetic element, called [Het-s]. [Het-s] is reversibly curable, the frequency of its appearance de novo is enhanced by overproduction of the protein encoded by the het-s gene, and the het-s gene is necessary for the propagation of [Het-s]. The protein encoded by the het-s gene is more protease-resistant in strains carrying [Het-s].
Prion variants
At this time, only a single [Het-s] variant is known, perhaps reflecting its being evolved to be a prion with specific properties, unlike the known yeast prions which appear to be diseases.
Species [Beta]
Host Saccharomyces cerevisiae
Distinguishing features
An enzyme that is necessary for its own activation (or maturation) will remain inactive if not initially activated or remain active in a self-propagating manner if initially active. This can be the basis for a prion (infectious protein) since transmission of the active enzyme from one individual to another lacking the active enzyme will change the self-propagating status of the recipient individual. In the absence of protease A, which normally can activate protease B, protease B acts as a prion of Saccharomyces cerevisiae.
Prion properties
Morphology
The beta prion is identical with the mature active form of the vacuolar protease B of yeast.
Physicochemical and physical properties
Active protease B is a soluble protein, synthesized as a precursor protein encoded by PRB1. Its activation requires both N- and C-terminal cleavage, which either protease A or mature protease B can carry out.
Proteins
The mature active form of Prb1p (vacuolar protease B) is a 284 aa residue protein which is derived by N- and C-terminal cleavages of a 635 residue precursor protein. Prb1p is a serine protease of the subtilisin group with roles in maturation of other vacuolar proteins, and in protein degradation by the vacuole which is critical for meiosis and spore formation and for survival of starvation.
Genome organization and replication
Normally, protease A, as well as mature protease B, can carry out the cleavage-maturation of proprotease B. In a pep4 deletion mutant lacking protease A, protease B acts as a prion. Cells initially lacking mature active protease B give rise to progeny, nearly all of which lack the active enzyme. Cells initially carrying active mature protease B give rise to offspring, nearly all of which also have active protease B. Transmission of active protease B to a cell lacking it, converts the pro-protease B in the recipient to the active form. This activity is then passed on to progeny by continued auto-activation. About 1 in 105 cells spontaneously develops protease B activity.
The protease B precursor protein requires removal of both N-terminal and C-terminal extension for maturation.
Antigenic properties
Not studied.
Biological properties
Like the amyloid-related prions, [BETA] is reversibly curable. Activity is lost by culturing cells on media that repress expression of Prb1p, but from a cured strain, [BETA] arises spontaneously in about 1 in 105 progeny cells. Overexpression of pro-Prb1p increases the frequency with which [BETA] arises about 100- to 1000-fold. The propagation of [BETA] requires the PRB1 gene, as expected. [BETA] is present in all wild-type cells, but is inapparent because protease A can activate proprotease B. Only in the absence of protease A does the prion character become evident.
[BETA] allows cells to go through meiosis and spore formation and to better survive starvation, a process that requires protein breakdown in the vacuole.
Prion variants
None reported.
Similarity with other taxa
The N-terminal prion domain of Sup35p includes repeat aa sequences similar to the octapeptide repeats in PrP, but the Sup35p repeats are not necessary for prion formation as shuffling the Sup35p prion domain does not prevent amyloid formation. The Sup35 prion domains of Candida albicans, Kluyveromyces lactis and Pichia methanolica can be prion domains in S. cerevisiae when fused to the S. cerevisiae Sup35MC (non-prion) domain. Full length Ure2p from S. bayanus, S. uvarum, S. mikatae, S. cariocanus and S. paradoxus can form the [URE3] prion in S. cerevisiae.
Overall, the yeast and fungal prions are analogous to the mammalian prion protein PrP, but not at all homologous.
Derivation of names
[BETA]: from protease B and the Greek letter beta.
[Het-s] and het-s: from heterokaryon incompatibility.
[PIN]: from [PSI] inducibility.
[PSI] from the greek letter Psi.
[URE3] and URE2: from ureidosuccinate.
[SWI]: from the Swi1 protein.
[OCT]: from the Cyc8 protein.
[MOT3]: from the Mot3 protein.
[ISP]: from the phenotype being the inverse of that or [PSI].
Prion: sigla from infectious protein.
Further reading
Chernoff et al., 1995 Y.O. Chernoff, S.L. Lindquist, B.-I. Ono, S.G. Inge-Vechtomov, S.W. Liebman, Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science. 268 (1995) 880–884.
Coustou et al., 1997 V. Coustou, C. Deleu, S. Saupe, J. Begueret, The protein product of the het-s heterokaryon incopatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl Acad. Sci., U S A. 94 (1997) 9773–9778.
Derkatch et al., 2001 I.L. Derkatch, M.E. Bradley, J.Y. Hong, S.W. Liebman, Prions affect the appearance of other prions: the story of [PIN]. Cell. 106 (2001) 171–182.
Edskes et al., 2009 H.K. Edskes, L.M. McCann, A.M. Hebert, R.B. Wickner, Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics. 181 (2009) 1159–1167.
King and Diaz-Avalos, 2004 C.Y. King, R. Diaz-Avalos, Protein-only transmission of three yeast prion strains. Nature. 428 (2004) 319–323.
Shewmaker et al., 2006 F. Shewmaker, R.B. Wickner, R. Tycko, Amyloid of the prion domain of Sup35p has an in-register parallel b-sheet structure. Proc. Natl Acad. Sci., U S A. 103 (2006) 19754–19759.
Tanaka et al., 2004 M. Tanaka, P. Chien, N. Naber, R. Cooke, J.S. Weissman, Conformational variations in an infectious protein determine prion strain differences. Nature. 428 (2004) 323–328.
Wickner, 1994 R.B. Wickner, Evidence for a prion analog in Saccharomyces cerevisiae: the [URE3] non-Mendelian genetic element as an altered URE2 protein. Science. 264 (1994) 566–569.
Wickner et al., 2004 R.B. Wickner, S.W. Liebman, S.J. Saupe, S.B. Prusiner, Prions of yeast and filamentous fungi: [URE3], [PSI+], [PIN+] and [Het-s]Prion Biology and Diseases. In: S.B. Prusiner, Prion Biology and Diseases. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY2004305–377.
Wickner et al., 2008 R.B. Wickner, F. Shewmaker, D. Kryndushkin, H.K. Edskes, Protein inheritance (prions) based on parallel in-register β-sheet amyloid structures. Bioessays. 30 (2008) 955–964.
Contributed by
Wickner, R.B.
Figures
Figure 1 Amyloid filaments of the Sup35 protein prion domain (courtesy of Dr Frank Shewmaker, NIH). Amyloid is a filamentous protein polymer, with strands perpendicular to the long axis of the filament, relative protease-resistance of the protein and special dye staining properties.
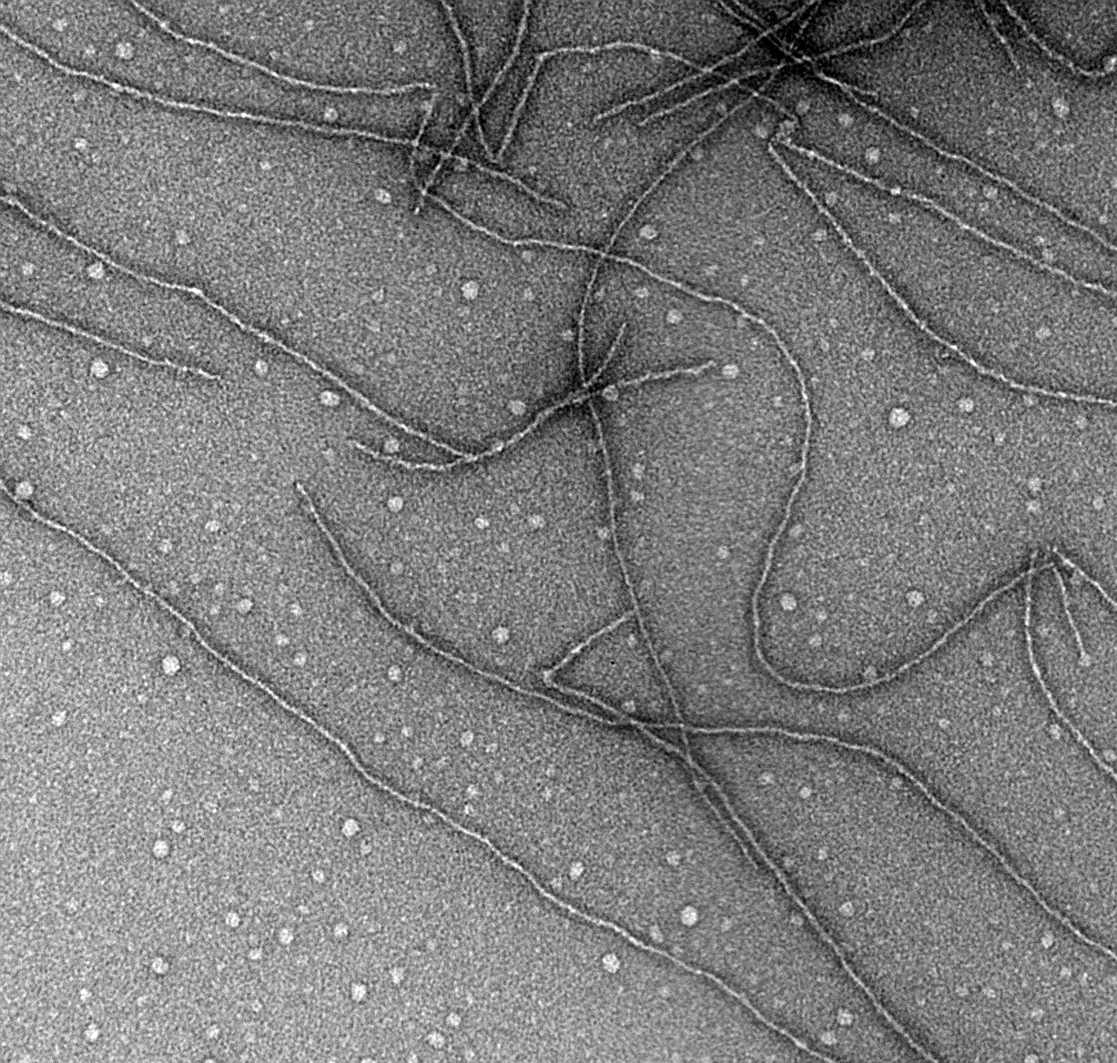
Figure 2 Architecture of yeast prion amyloid filaments is an in-register parallel -sheet. The side chains of each residue form a row along the long axis of the filaments, and interactions among these identical side chains, such as the -zipper of glutamine or asparagine side chains, enforce the in-register structure. The same interactions explain how conformational information is propagated from the filament to new molecules joining the ends.
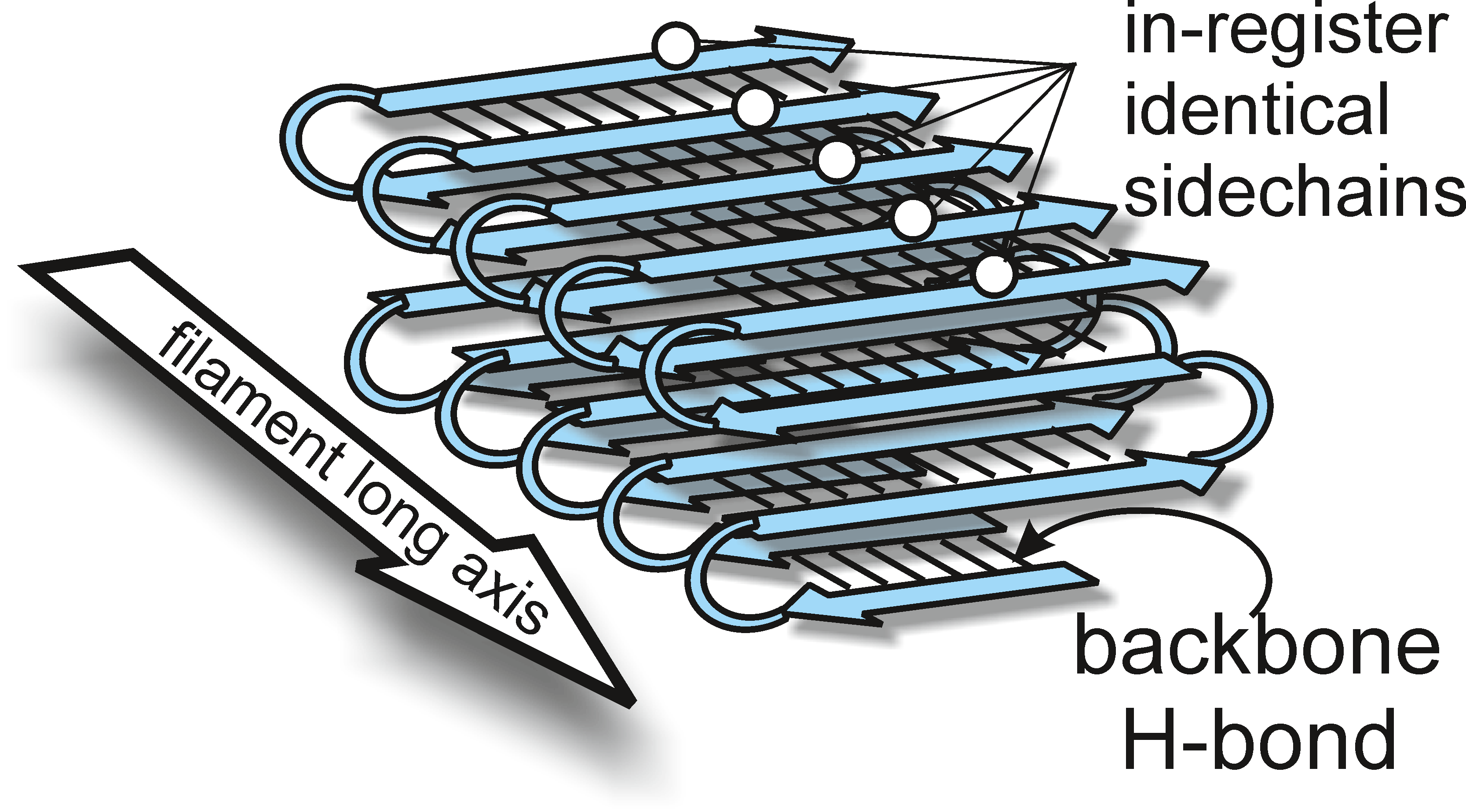
Figure 3 Amyloid prions replicate by elongation of amyloid filaments and scission of filaments by the action of chaperones to produce new filaments.
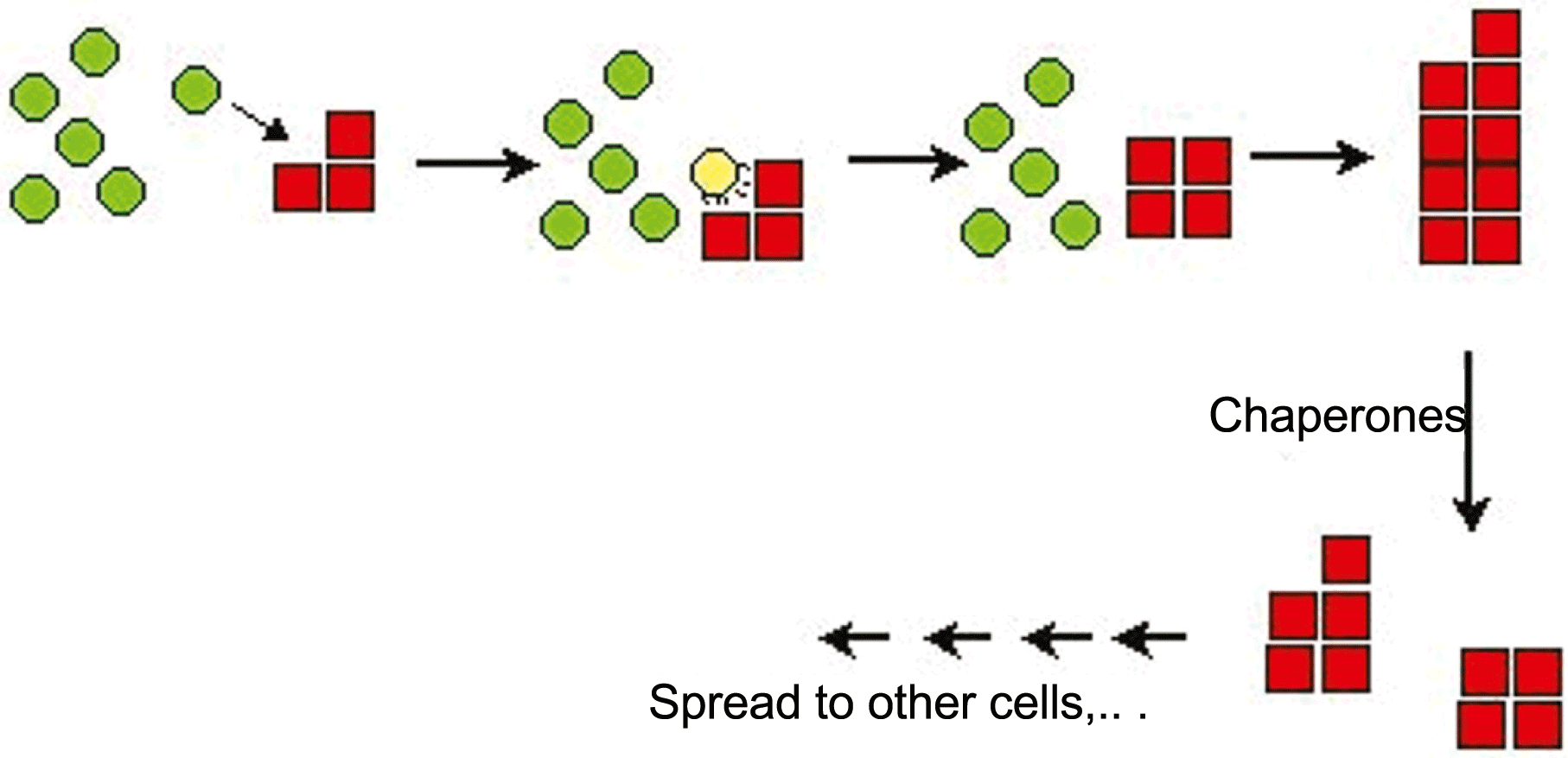
Figure 4 Genetic criteria for identification of prions.
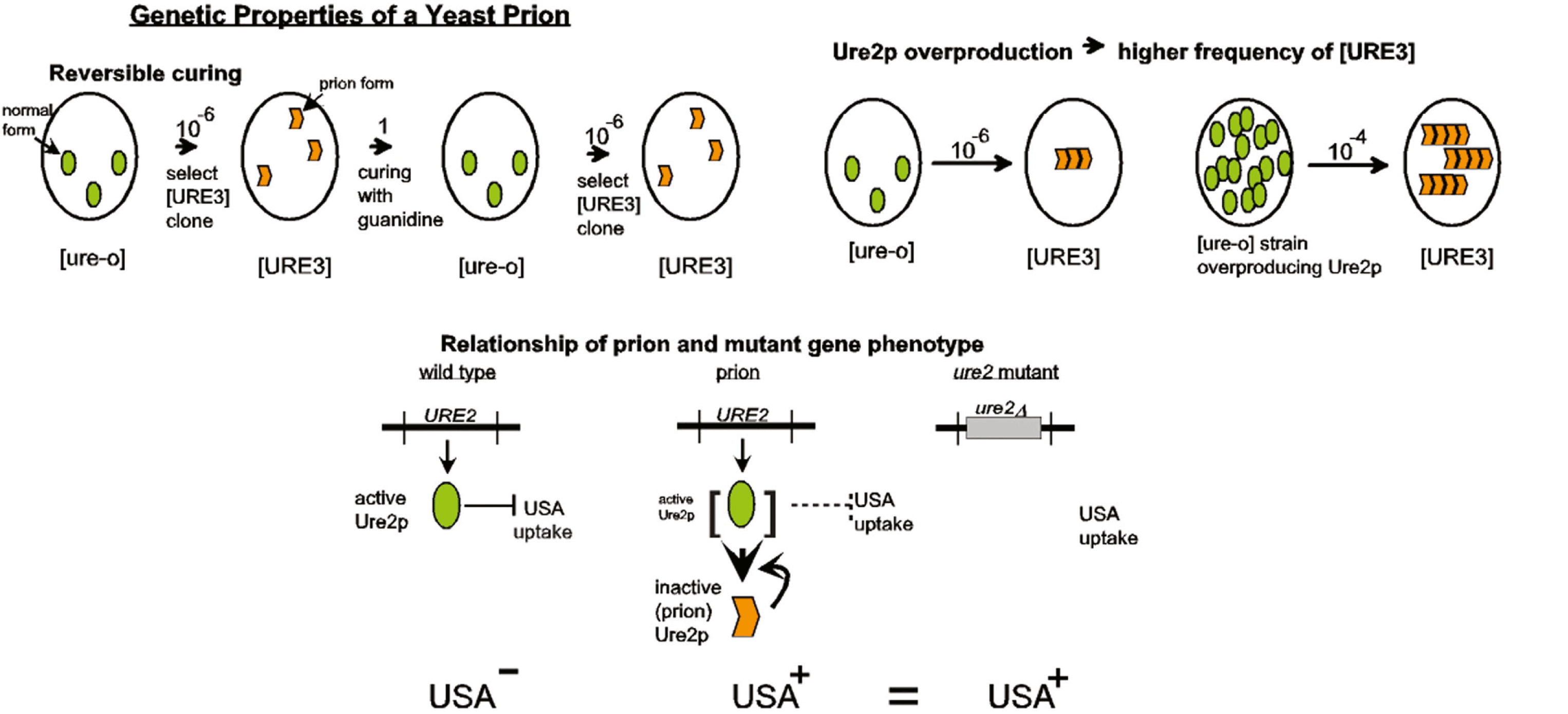
Figure 5 Amyloid filaments in cells of S. cerevisiae infected with the [URE3] prion.
(From Speransky, V. et al. (2001). Prion filament networks in [URE3] cells if Saccaromyces cerevisiae. J. Cell Biol., 153, 1327-1335; with permission.)