Family: Parvoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Parvoviruses are among the smallest of isometric viruses, with diameters ranging from 215 Å (Penaeus stylirostris densovirus, PstDNV) to 255 Å (canine parvovirus, CPV). X-ray crystallography studies have unequivocally established the icosahedral nature of parvoviruses, with 31 rotational elements (six 5-folds, ten 3-folds and fifteen 2-folds). The final resolution obtained ranges from 3.7 (Galleria mellonella densovirus; GmDNV) to 2.5 (PstDNV) Å, but the N-termini of the capsid proteins have not yet been solved. Some structures have a smooth appearance (GmDNV), whereas others (adeno-associated virus-2, AAV-2, and PstDNV) are spiky at the 3- or 5-fold symmetry axes (Figure 1). Parvoviruses lack envelopes.
Physicochemical and physical properties
Infectious particles are composed of about 75% protein and about 25% DNA, and their Mr is about 5.5–6.2 ×106. Virion buoyant density is 1.39–1.43 g cm−3 in CsCl and the S20,w is 110–122S. Infectious particles with buoyant densities of about 1.45 g cm−3 may represent conformational or other variants, or precursors to the mature particles. Defective particles with deletions in the genome occur and exhibit lower densities (about 1.34 g cm−3). Mature virions are stable in the presence of lipid solvents, or on exposure to pH 3–9 or, for most species, incubation at 56 °C for 60 min. Viruses can be inactivated by treatment with formalin, β-propriolactone, hydroxylamine, ultraviolet light and oxidizing agents such as sodium hypochlorite, although virus aggregates are somewhat more resistant.
Nucleic acid
The genome is a linear, nonsegmented molecule of ssDNA, 4–6.3 kb in size (Mr 1.5–2.0 ×106), with a G+C content of about 40–55%. The 5′- and 3′-ends of the genome contain palindromic sequences, 120 to ∼550 nt in length, which can be folded into hairpin structures essential for viral DNA replication. These terminal hairpins may be part of a terminal repeat, and therefore related in sequence (e.g. dependoviruses), whereas some genomes have terminal hairpins that are unrelated in sequence to one another (e.g. members of the genus Parvovirus). Some parvoviruses preferentially encapsidate ssDNA of negative polarity (i.e. complementary to the viral mRNA species; e.g. minute virus of mice, MVM), whereas others may encapsidate ssDNA species of either polarity in equivalent (e.g. adeno-associated viruses, AAVs) or different proportions (e.g. bovine parvovirus, BPV). The percentage of particles encapsidating the positive strand can vary from 1 to 50% and may be influenced by the host cell in which the virus is produced (e.g. LuIII virus, LuIIIV). Some insect parvoviruses (densoviruses) have genes on both strands.
Proteins
Most parvoviruses have 2–4 virion proteins, and up to five in the case of brevidensoviruses and pefudensoviruses (VP1-VP5). Depending on the species, protein sizes are: VP1 75–96 kDa, VP2 65–85 kDa, VP3 55–75 kDa and VP4 45–52 kDa (for brevidensoviruses somewhat smaller). The viral proteins represent different N-extended forms of the same gene product, except for the pefudensoviruses that generate two different N-terminal extensions. Some shrimp densoviruses have a single 54 kDa capsid protein. The VP1-specific regions of viruses of all genera, except members of the genera Brevidensovirus and Amdovirus and some unclassified parvoviruses, contain the enzymatic core of a phospholipase A2 (PLA2). The two smallest proteins are usually the principal proteins. Spermidine, spermine and putrescine have been identified as components of some densoviruses and iteraviruses.
Lipids
Virions lack essential lipids.
Carbohydrates
None of the viral proteins are known to be glycosylated.
Genome organization and replication
Parvoviruses usually possess two major gene cassettes (Figure 2). The REP ORF encodes the non-structural proteins (NS), which are required for transcription and DNA replication, and the CP ORF encodes the structural proteins of the capsid (the CAP, VP or S proteins). Both gene cassettes are present on the same DNA strand of all parvoviruses except for members of the genera Densovirus and Pefudensovirus and some as yet unclassified ambisense densoviruses. The REP functions and the CPs of the latter genera are encoded in the 5’-halves of the complementary strands and represented, as for monosense parvoviruses, with the REP cassette to the left (Figure 2). In some viruses, other minor ORFs have been detected. For some of these, a protein product has been identified (the ORF for the N terminus of VP1). Mutations within the REP ORF block virus replication and gene expression. Mutants in REP or CP can be complemented in trans. The palindromic sequences (at both termini) are required in cis for DNA replication to occur.
Some viruses use alternative splicing, leaky scanning, or a combination thereof, to obtain different forms of the REP and CAP gene products. The MVM REP ORF produces two major non-structural proteins NS1 and NS2P, and two minor ones (NS2Y and NS2L). A subset of the same alternative splicing strategy allows translation of the CP ORF to produce two proteins, VP1 and VP2. MVM VP3 is generated in the intact virion by proteolytic cleavage of VP2. VP1 and VP2 are identical except for their N termini. Synthesis of VP1 derives from a minor spliced mRNA containing a methionine initiation codon that allows translation of a small ORF, which encodes basic amino acid sequence motifs, upstream (5′) of the VP2-coding sequence. This expression strategy varies among viruses in the different genera and even, to a minor extent, within a genus. Parvoviruses use an alternative splice donor, while dependoviruses use an alternative splice acceptor for this purpose. The use of a leaky scanning mechanism for the generation of NS and VP products is common among densoviruses.
Members of the genera Erythrovirus, Amdovirus and Bocavirus use only a single promoter in their left genome end, and so regulation of their expression must be exclusively post-transcriptional. All of the parvoviruses make extensive use of alternative splicing strategies, and all members of the genera Erythroviruses, Dependovirus, Amdovirus and Bocavirus examined also use alternative polyadenylation at a site within the centre of the genome. In addition, the parvoviruses make use of alternative translation strategies for the expression of both their structural and nonstructural proteins: regulation of translation is known to govern the expression of the VP2 protein of AAV-2, the capsid proteins generated by members of the genera Amdovirus and Bocavirus, and the nonstructural proteins of the goose parvovirus (GPV) subgroup of the genus Dependovirus.
Other parvoviruses contain two (genera Parvovirus and Densovirus) or three (genera Dependovirus and Brevidensovirus) promoters for mRNA transcription. Some of the mRNAs are spliced, thus allowing alternative forms of the protein products to be produced. The mRNA species are capped and polyadenylated either at a common 3’ site near the end of the genome (MVM and AAVs of primates), or at an alternative polyadenylation site in the centre of the genome as well as at a site near the end of the genome (all known members of the genus Dependovirus other than those that infect primates). Depending on the species, viruses may benefit from co-infection with other viruses, such as adenoviruses or herpesviruses, or from the effects of chemical or other treatments of the host cells. Viral proteins accumulate in the nucleus in the form of empty capsid structures. Progeny infectious virions accumulate in the cell nucleus.
Viral entry into the cell is usually by receptor-mediated endocytosis and is blocked by antagonists of vacuolar ATPase. The trafficking of virus within the cell appears to vary among members of the family, and even among species within individual genera. The PLA2 domain plays a role in the trafficking or release of particles from endosomal compartments. The process of uncoating is not well understood. Virus replication takes place in the cell nucleus and appears to require the cell to go through S-phase, indicating a close association between the host and virus replication processes. Autonomously replicating parvoviruses probably do not initiate gene expression until the host cell enters S-phase to produce viral dsDNA, whereas this transition is effected by a positive process under the control of the helper virus in the case of helper-dependent parvoviruses.
Replication proceeds through a series of duplex, concatemeric intermediates by the rolling hairpin mechanism (Figure 3), and probably involves a processive host DNA polymerase(s) (probably pol δ, possibly pol or others). In step (a) the base-paired 3’ nucleotide of the left-end hairpin is used by a host polymerase to prime conversion of virion DNA to the first duplex intermediate. This generates a monomer length duplex molecule in which the two strands are covalently continuous at the viral left-end telomere. Synthesis of this intermediate precedes viral gene expression. The 3’ end of the new DNA strand is ligated to the 5’ end of the hairpin by a host ligase, creating a covalently continuous duplex molecule (step b). Replication beyond this point requires expression of NS1, which carries out a “hairpin transfer” reaction, in which it nicks the ligated strand (step c). The replication fork now unfolds and copies the hairpin, thus replacing the original sequence of the terminus with its inverted complement (step d). The terminal sequences are imperfect palindromes, and since this inversion occurs with every round of replication, progeny genomes comprise equal numbers of each terminal orientation, dubbed “flip” and “flop”. In MVM replication, this hairpin transfer reaction occurs only at the right-end, because of different structural and co-factor requirements needed to activate the NS1 nickase at each terminus.
When inversion occurs on the first monomer formed after uncoating, it regenerates the “tether” sequence, lost during entry, and now attached to a newly synthesized NS1 molecule. Extended-form right-end termini are melted out and reformed into hairpin “rabbit ear” structures in a process facilitated by direct binding of NS1 to sequences in the terminus (step e). This allows the newly synthesized DNA to create the base-paired hairpin structures needed to prime synthesis of additional linear sequences (step f). This gives rise to a palindromic duplex dimeric (step g), which can undergo the same right-end rearrangement (step h), leading to the synthesis of tetrameric concatemers (step h), in which alternating unit length genomes are fused in left-end:left-end and right-end:right-end orientations. Individual genomic monomer duplexes are then excised from these concatemers by a process called junction resolution.
Antigenic properties
Parvoviruses appear to have very stable virions that are quite simple antigenically. This has led to the use of individual serotype as a major criterion for species demarcation. Serotype has been defined by neutralization of infectivity in cell culture, hemagglutination-inhibition or specific ELISA using a capture format. Two antigenic sites, defined by mutations that confer resistance to neutralization by monoclonal antibodies, have been determined for CPV. Some, but not all, viruses representing species in a genus may be antigenically related by epitopes in the NS proteins.
Biological properties
Autonomous parvoviruses require host cell passage through S-phase. Certain parvoviruses replicate efficiently only in the presence of helper viruses (e.g. adenoviruses or herpesviruses). These helper functions involve the adenovirus or herpesvirus early gene products and trans-activation of parvovirus replication. The helper functions appear to relate to effects of the helper virus upon the host cell rather than direct involvement of helper virus gene products in parvovirus replication. Association of parvoviruses with tumor cell lines appears to relate to increased DNA replication and/or the state of differentiation in such cells, rather than previous involvement as an etiologic agent of oncogenesis. Co-infection involving certain parvoviruses and selected oncogenic adenoviruses (or other viruses) may reduce the oncogenic effect of those viruses, possibly by promoting cell death. In certain circumstances, parvovirus DNA may integrate into the host genome, from which it may be activated by subsequent helper virus infection. The site of integration may be specific in certain hosts (e.g. the q arm of human chromosome 19 for AAV-2).
Subfamily Parvovirinae
Distinguishing features
Viruses assigned to the subfamily Parvovirinae infect vertebrates and vertebrate cell cultures, sometimes in association with other viruses.
Genus Parvovirus
Type species Minute virus of mice
Distinguishing features
For some members of the genus, mature virions contain virtually only negative strand DNA of 5 kb. In other members, positive strand DNA occurs also in variable proportions (1–50%). The linear molecule of ssDNA has hairpin structures at both the 5’ and 3’ ends. The 3’ terminal hairpin (left end, “–” strand) is 115–116 nt in length, the 5’ structure is 200–242 nt long. There are two mRNA promoters (map units 4 and 39) and a single polyadenylation site at the 3’ end. Characteristic cytopathic effects are induced by the viruses during replication in cell culture. Many species exhibit hemagglutination with red blood cells of one or more species. Under experimental conditions, the host range may be extended to a large number of vertebrate species (e.g. rodent viruses and LuIII virus (LuIIIV) replicate in Syrian hamsters). Transplacental transmission has been detected for a number of species.
Genome organization and replication
Alternate splicing controls viral gene expression (Figure 4). For MVM, transcripts 1–6 are made from the promoter at 4 map units. Transcripts 1–3 encode only NS1, while transcripts 4–6, which are spliced into another reading frame by the major intron, encode NS2P, NS2Y and NS2L, respectively, the C-termini of which are different due to the use of alternative donor and acceptor splice sites bordering the small intron. A promoter at 38 map units is transactivated by NS1 to drive the synthesis of transcripts 7–9. The use of the same alternative donor splice sites that are present in the P4 transcripts controls the synthesis of the CPs, such that transcripts 7 and 9 encode VP2, while transcript 8 encodes VP1.
Species demarcation criteria in the genus
Members of each species are antigenically distinct, as assessed by neutralization using polyclonal antisera, and natural infection is usually confined to a single host species. Generally, species are <95% related by non-structural gene DNA sequence.
List of species in the genus Parvovirus
| Chicken parvovirus1 |
|
|
| Chicken parvovirus | [GU214704] | (ChPV) |
| Feline panleukopenia virus |
|
|
| Canine parvovirus strain CPV-N | [M19296] | (CPV) |
| Feline panleukopenia virus-b | [M75728] | (FPV-b) |
| Mink enteritis virus Abashiri | [D00765] | (MEV) |
| Raccoon parvovirus Georgia | [M24005] | (RPV) |
| H-1 parvovirus |
|
|
| H-1 parvovirus | [X01457] | (H-1PV) |
| HB parvovirus |
|
|
| HB parvovirus |
| (HBPV) |
| Kilham rat virus |
|
|
| Kilham rat virus | [AF321230] | (KRV) |
| Lapine parvovirus |
|
|
| Lapine parvovirus |
| (LPV) |
| LuIII virus |
|
|
| LuIII virus | [M81888] | (LuIIIV) |
| Minute virus of mice |
|
|
| Minute virus of mice (Cutter) | [U34256] | (MVMc) |
| Minute virus of mice (immunosuppressive) | [M12032] | (MVMi) |
| Minute virus of mice (prototype) | [J02275] | (MVMp) |
| Mouse parvovirus 1 |
|
|
| Mouse parvovirus 1 | [U12469] | (MPV-1) |
| Porcine parvovirus |
|
|
| Porcine parvovirus Kresse | [U44978] | (PPV-Kr) |
| Porcine parvovirus NADL-2 | [L23427] | (PPV-NADL2) |
| RT parvovirus |
|
|
| Rat parvovirus 1 | [AF036710] | (RTPV-1) |
| Tumor virus X |
|
|
| Tumor virus X |
| (TVX) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
1 A proposal to reclassify the species Chicken parvovirus is currently under consideration by the ICTV.
List of other related viruses which may be members of the genus Parvovirus but have not been approved as species
| Hamster parvovirus | [U34255] | (HaPV) |
| Mouse parvovirus 2 |
| (MPV-2) |
| Rat minute virus 1 | [AF332882] | (RMV-1) |
| Tumor virus X |
| (TVX) |
Genus Erythrovirus
Type species Human parvovirus B19
Distinguishing features
Populations of mature virions contain equivalent numbers of positive and negative sense ssDNA, 5.5 kb in size. The DNA molecules contain inverted terminal repeats of 383 nt, the first 365 nt of which form a palindromic sequence. Upon extraction, the complementary DNA strands usually self-anneal to form dsDNA. There is a single mRNA promoter (map unit 6) and two polyadenylation signals: one near the middle of the genome, the other near the 3’ end. Efficient replication occurs in primary erythrocyte precursors. There have also been reports of productive infection in cell lines of megakaryoblastoid erythroleukemic origin.
Genome organization and replication
For human parvovirus B19 (B19V) there is only one promoter, at 6 map units, but two alternative polyadenylation sites (Figure 5). Transcripts 1 and 2 encode VP1, and transcript 3 encodes NS1. Two small ORFs can also be accessed by these alternatively spliced mRNAs, depending upon the relative strength of the initiation codons. Transcripts 8 and 9 encode an 11 kDa protein containing three proline-rich regions that conform to consensus Src homology 3 (SH3) ligand sequences. Transcripts 1, 4, 6 and 8 are predicted to translate a 7.5 kDa polypeptide of unknown function.
Biological properties
B19V causes Fifth Disease, polyarthropathia, anemic crises in children with underlying hematological diseases (e.g. sickle cell anemia or thalassemia) and intra-uterine infections (with hydrops fetalis in some cases).
Species demarcation criteria in the genus
Members of each species are probably antigenically distinct, and natural infection is confined to a single host species. Species are <95% related by non-structural gene DNA sequence.
List of species in the genus Erythrovirus
| Human parvovirus B19 |
|
|
| Human parvovirus B19-A6 | [AY064475, AY064476] | (B19V-A6) |
| Human parvovirus B19-Au | [M13178] | (B19V-Au) |
| Human parvovirus B19-lali | [AY044266] | (B19V-LaLi) |
| Human parvovirus B19-V9 | [AJ223617, AJ242810] | (B19V-V9) |
| Human parvovirus B19-Wi | [M24682] | (B19V-Wi) |
| Human parvovirus B19-J35 | [AY386330] | (B19V-J35) |
| Pig-tailed macaque parvovirus |
|
|
| Pig-tailed macaque parvovirus | [AF221123] | (PmPV) |
| Rhesus macaque parvovirus |
|
|
| Rhesus macaque parvovirus | [AF221122] | (RmPV) |
| Simian parvovirus |
|
|
| Simian parvovirus (cynomolgus) | [U26342] | (SPV) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Erythrovirus but have not been approved yet as species
| Chipmunk parvovirus | [AF406967] | (ChpPV) |
Genus Dependovirus
Type species Adeno-associated virus-2
Distinguishing features
Populations of mature virions contain equivalent numbers of positive or negative strand ssDNA about 4.7 kb in size. The DNA molecules contain inverted terminal repeats of 145 nt, the first 125 nt of which form a palindromic sequence. Upon extraction, the complementary DNA strands usually form dsDNA. There are three mRNA promoters (map units 5, 19, 40) (Figure 6). For all currently accepted members of the genus Dependovirus, except for the duck and goose parvoviruses, efficient replication is dependent upon helper adenoviruses or herpesviruses. Under certain conditions (presence of mutagens or synchronization of cell replication with hydroxyurea), replication can also be detected in the absence of helper viruses. All isolates of adeno-associated virus share a common antigen as demonstrated by fluorescent antibody staining. Transplacental transmission has been observed for adeno-associated virus-1 (AAV-1) and vertical transmission has been reported for avian adeno-associated virus (AAAV).
Genome organization and replication
Dependoviruses have three transcriptional promoters, at 5, 19 and 40 map units, which transcribe mRNAs in a temporally regulated fashion throughout infection (Figure 6). P5 transcripts are the first to be expressed, followed by those from P19, then those from P40. P5 transcript 1 encodes the non-structural proteins Rep78, and transcripts 2 and 3 encode Rep 68. These two forms of the Rep protein differ at their C-termini. Likewise, P19 transcripts encode two Rep forms, Rep52 from transcript 4 and Rep48 from trancripts 5 and 6. P40 transcripts encode the structural proteins, transcript 8 encoding VP1 and transcript 9 encoding VP2 and VP3 by an alternate translation initiation mechanism. The unspliced P40 transcript (7) does not appear to encode a functional protein.
RNAs generated from both the P7 (analogous to AAV-2 P5) and P19 promoters of all the non-primate members of the genus Dependovirus (exemplified by AAV-5 in Figure 6) are polyadenylated efficiently at a site lying within the intronic region in the centre of the genome, termed (pA)p. Because AAV5 P7- and P19-generated transcripts are polyadenylated at this site and not spliced, Rep78 and Rep52 are the only Rep proteins expressed as primary translation products during AAV-5 infection.
Species demarcation criteria in the genus
Members of each species are antigenically distinct, as assessed by neutralization using polyclonal antisera, and natural infection is usually confined to a single host species. Generally, species are <95% related by non-structural gene DNA sequence.
List of species in the genus Dependovirus
| Adeno-associated virus-1 |
|
|
| Adeno-associated virus-1 | [AF063497] | (AAV-1) |
| Adeno-associated virus-6 | [AF208704] | (AAV-6) |
| Adeno-associated virus-2 |
|
|
| Adeno-associated virus-2 | [J01901] | (AAV-2) |
| Adeno-associated virus-3 |
|
|
| Adeno-associated virus-3 | [AF028705] | (AAV-3) |
| Adeno-associated virus-4 |
|
|
| Adeno-associated virus-4 | [U89790] | (AAV-4) |
| Adeno-associated virus-5 |
|
|
| Adeno-associated virus-5 | [AF085716] | (AAV-5) |
| Avian adeno-associated virus |
|
|
| Avian adeno-associated virus | [AY186198] | (AAAV) |
| Bovine adeno-associated virus |
|
|
| Bovine adeno-associated virus |
| (BAAV) |
| Canine adeno-associated virus |
|
|
| Canine adeno-associated virus |
| (CAAV) |
| Duck parvovirus |
|
|
| Duck parvovirus | [U22967] | (BDPV) |
| Equine adeno-associated virus |
|
|
| Equine adeno-associated virus |
| (EAAV) |
| Goose parvovirus |
|
|
| Goose parvovirus | [U25749] | (GPV) |
| Ovine adeno-associated virus |
|
|
| Ovine adeno-associated virus |
| (OAAV) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Dependovirus but have not been approved as species
| Adeno-associated virus-7 | [AF513851] | (AAV-7) |
| Adeno-associated virus-8 | [AF513852] | (AAV-8) |
| Adeno-associated virus-9 | [AX753250] | (AAV-9) |
| Snake adeno-associated virus | [AY349010] | (SAAV) |
Genus Amdovirus
Type species Aleutian mink disease virus
Distinguishing features
Most features are shared with members of the genera Parvovirus and Bocavirus. Mature virions contain negative strand DNA of 4748 nt in length. Permissive replication is observed only in Crandell feline kidney cells, although restricted replication is observed, and may be antibody-dependent, in cells bearing Fc receptors (e.g. macrophages). Evidence of infection has been detected in most mustelids, skunks and raccoons. Virion structure differs slightly from members of the genera Parvovirus and Bocavirus, and resembles that of the genus Dependovirus. The primary difference is the presence of three mounds elevated above the capsid surface around the three-fold icosahedral axis of symmetry, similar to those observed for dependovirus virions. The VP1 N terminus is much shorter than those found for other members of the subfamily Parvovirinae, and there is no evidence of a phospholipase 2A enzymatic core.
Genome organization and replication
AMDV generates six species of mRNAs, which are produced by alternative splicing and alternative polyadenylation of a pre-mRNA generated by a single promoter at the left end of the genome (Figure 7). Three different splicing patterns are used, and each type is found polyadenylated at either the distal 3’ end of the genome or at a proximal site (pA)p in the centre of the genome. The R1 class of mRNAs encode the viral replicator protein NS1. The R2 mRNA, which is the predominant RNA produced during infection, expresses the NS2 coding region and is also essentially the sole source of the viral capsid proteins VP1 and VP2. Caspase processing of the NS1 protein is required to produce a sub-species that locates to the nucleus. This processing is essential for completion of the viral life cycle.
Antigenic properties
All isolates in the species appear to be antigenically indistinguishable.
Biological properties
In susceptible adult hosts, pathogenic isolates cause a persistent, restricted infection associated with a progressive disorder of the immune system, including plasmacytosis, glomerulonephritis and hypergammaglobulinemia. Extremely high levels of antiviral antibody are directed at determinants on the virus capsid surface. In newborn animals, infection is permissive and causes a fulminant interstitial pneumonitis that is often fatal. Survivors develop an adult form of disease.
List of species in the genus Amdovirus
| Aleutian mink disease virus |
|
|
| Aleutian mink disease virus-G | [M20036] | (AMDV-G) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
Genus Bocavirus
Type species Bovine parvovirus
Distinguishing features
The genome is ssDNA of 5.5 kb, with a monosense strategy and non-identical terminal palindromes. Sequence analysis of BPV and canine minute virus (CnMV) demonstrate that, while their NS1 and VP1 genes are 34% and 41% similar to one another, the two genomes are very distinct from all other clusters of viruses in the subfamily Parvovirinae. In addition, the bocavirus genome encodes a 22.5 kDa nuclear phosphoprotein, NP-1, that is distinct from any other parvovirus-encoded polypeptide.
Genome organization and replication
Large ORFs within the left and right halves of the genome encode the NS and VP proteins, respectively, while a shorter ORF in the middle of the genome, overlapping the C-terminal sequence of the NS1 protein, encodes NP-1. All RNAs are generated from a single promoter at the left genome end at approximately 4.5 mu. BPV RNAs are alternatively spliced and alternatively polyadenylated (either at a site in the centre or at the 3’-end of the genome) to generate at least 8 mRNAs (Figure 8).
Species demarcation criteria in the genus
Members of each species are probably antigenically distinct, and natural infection is confined to a single host species. Species are <95% related by non-structural gene DNA sequence. The four distinct genetic lineages of human bocavirus show a significant divergence, >8% amino acid and >10% nucleotide difference in the complete VP gene, but a low level of intraspecies diversity.
List of species in the genus Bocavirus
| Bovine parvovirus |
|
|
| Bovine parvovirus | [M14363] | (BPV) |
| Canine minute virus |
|
|
| Canine minute virus | [AF495467] | (CnMV) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the subfamily Parvovirinae but have not yet been approved as a genus or a species
Human bocavirus 1 (HBoV-1) is primarily a respiratory virus, whereas human bocavirus 2 to 4 (HBoV-2, HBoV-3 and HBoV-4) are prevalent in stool samples.
| Human bocavirus-1 | [DQ000496] | (HBoV-1) |
| Human bocavirus-2 | [FJ170278] | (HBoV-2) |
| Human bocavirus-3 | [EU918736] | (HBoV-3) |
| Human bocavirus-4 | [FJ973561] | (HBoV-4) |
Unclassified vertebrate parvoviruses
Among vertebrate parvoviruses that may need to be reclassified into new genera or have not yet been classified, three groups can be discerned: PARV4 and related viruses, chicken parvovirus and related viruses, and a diverse group with no clear taxonomical structure. These groups lack significant sequence identity with members of the accepted genera in the subfamily Parvovirinae, and are also significantly different from each other.
PARV4 virus (PARV4) is widely distributed in injecting drug users in the United States and Europe. Genotypes 1 and 2 (formerly named PARV5, found in older coagulation concentrates) are phylogenetically distinct, as are PARV4 sequences detected in a study of sub-Saharan Africans (93%; equidistant from genotypes 1 and 2). Additionally, PARV4-like viruses, with a 60–65% nucleotide identity but identical genome organizations, were recently identified at high frequencies in porcine and bovine tissue samples. These viruses have a 5.3 kb genome and ITRs of about 150–200 nt. The largest capsid protein contains also a PLA2 motif and a conserved SAT sequence that may be expressed late during infection. A new genus has been proposed for this group.
| Bovine hokovirus | [EU200669] | (BoHV) |
| PARV4 virus | [AY622943] | (PARV4V) |
| Porcine hokovirus | [EU200677] | (PoHV) |
The second group contains chicken and turkey parvoviruses, which have a 5 kb genome with a somewhat larger ITR (about 210 nt). However, the capsid proteins do not contain the PLA2 domain that most parvoviruses use for cell entry. A new genus has been proposed for this group.
| Turkey parvovirus 260 | [GU214706] | (TuPV-260) |
Several parvoviruses from bovine and porcine tissues remain unclassified. Erythroviruses are the closest relatives of bovine parvovirus 3 (BPV3), with a 5.3 kb genome, but remain quite different. Bovine parvovirus 2 (BPV2) has a 5.6 kb genome with two ORFs (phylogenetically different from any other parvovirus). Porcine parvovirus 4 (PPV4) is closest to BPV2 but has an additional ORF in the middle of the genome. The only known porcine parvovirus 2 (PPV2) sequence is part of a dimeric replicative form. These unassigned viruses all contain a PLA2 domain.
| Bovine parvovirus-2 | [[AF406966] | (BPV2) |
| Bovine parvovirus-3 | [AF406967] | (BPV3) |
| Porcine parvovirus-2 | [AB076669] | (PPV2) |
| Porcine parvovirus-4 | [GQ387499] | (PPV4) |
Subfamily Densovirinae
Distinguishing features
Viruses assigned to the subfamily Densovirinae infect arthropods. The ssDNA genome of virions is either monosense (viruses in the genera Iteravirus and Brevidensovirus, and some unassigned densoviruses) or ambisense (viruses in the genera Densovirus and Pefudensovirus, and some unassigned densoviruses). Either strand may be packaged. There may be 1–5 structural proteins. Viruses multiply efficiently either in most of the tissues of larvae, nymphs and adults of the host species, without the involvement of helper viruses, or exclusively in the midgut. Cellular changes consist of hypertrophy of the nucleus with accumulation of virions therein to form dense, voluminous, intranuclear masses. The known host range includes members of the insect orders Dictyoptera, Diptera, Hemiptera, Homoptera, Lepidoptera, Odonata and Orthoptera. Some densoviruses infect and multiply in shrimps.
Genus Iteravirus
Type species Bombyx mori densovirus
Distinguishing features
The ssDNA genome is about 5 kb in size with ITRs. Populations of virions encapsidate equal numbers of plus and minus strands. ORFs for both the structural and nonstructural proteins are located on the same strand.
Genome organization and replication
There is apparently one mRNA promoter upstream of each ORF. There is a small ORF on the complementary strand of some of these viruses that may not be coding. The DNA has an inverted terminal repeat of 230 nt, the first 159 nt are palindromic and can form a J-shaped hairpin structure when folded. There are two ORFs for non-structural proteins and one ORF for four structural proteins.
Species demarcation criteria in the genus
Members of each species are probably antigenically distinct, and natural infection is confined to a single host species. Species are <95% related by non-structural gene DNA sequence.
List of species in the genus Iteravirus
| Bombyx mori densovirus |
|
|
| Bombyx mori densovirus | [AY033435] | (BmDNV) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Iteravirus but have not been approved as species
| Casphalia extranea densovirus | [AF375296] | (CeDNV) |
| Dendrolimus punctatus densovirus | [AY665654] | (DpDNV) |
| Sibine fusca densovirus |
| (SfDNV) |
Genus Brevidensovirus
Type species Aedes aegypti densovirus
Distinguishing features
The genome is about 4 kb in size with terminal hairpins, but no ITRs. ORFs for the structural and nonstructural proteins are on the same strand. The brevidensoviruses are at least as different, in sequence, from other members of the subfamily Densovirinae as these are from any member of the subfamily Parvovirinae. Populations of virions encapsidate positive and negative strands, but a majority of strands are of negative polarity (85%).
Genome organization and replication
A palindromic sequence of 146 nt is at the 3′-end of the genome and a different palindromic sequence of 164 nt is at the 5′-end. Both terminal sequences can fold to form a T-shaped structure. The genome does not contain any sequence recognizable as a PLA2 domain. There are mRNA promoters at map units 7, 7.5 and 60. There is a small ORF of unknown function on the complementary strand. PstDNV, infecting shrimps, has a weak sequence identity with other brevidensoviruses, probably partially because of different codon preferences of the hosts. However, the overall genome organization is similar.
Species demarcation criteria in the genus
Members of each species are probably antigenically distinct, and natural infection is confined to a single host species. Species are <95% related by non-structural gene DNA sequence.
List of species in the genus Brevidensovirus
| Aedes aegypti densovirus |
|
|
| Aedes aegypti densovirus | [AY160976] | (AaeDNV) |
| Aedes albopictus densovirus |
|
|
| Aedes albopictus densovirus | [AY095351] | (AalDNV) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Brevidensovirus but have not been approved as species
| Anopheles gambiae densovirus | [EU233812] | (AgDNV) |
| Culex pipiens pallens densovirus | [EF579756] | (CppDNV) |
| Haemogogus equinus densovirus | [AY605055] | (HeDNV) |
| Penaeus stylirostris densovirus | [AF273215] | (PstDNV) |
| Simulium vittatum densovirus |
| (SvDNV) |
| Toxorhynchites amboinensis densovirus |
| (TaDNV) |
| Toxorhynchites splendens densovirus | [AF395903] | (TsDNV) |
Genus Densovirus
Type species Junonia coenia densovirus
Distinguishing features
The ssDNA genome is about 6 kb in size with long (>0.5 kb) inverted terminal repeats and ambisense organization (Figure 2). Populations of virions encapsidate equal amounts of positive and negative strands.
Genome organization and replication
On one strand there are three ORFs which encode NS proteins on mRNAs transcribed from a promoter 10 map units from the left end. The four structural proteins are encoded on the complementary strand, on an mRNA transcribed from a promoter at 90 map units, towards the centre of the genome. The NS and VP transcripts have a short overlap in the centre of the genome. NS3 protein is produced by an unspliced mRNA, whereas NS1 and NS2 are produced by a spliced mRNA and leaky scanning translation. The four VP proteins (VP1-4), and NS1 and NS2 are produced by a leaky scanning translation initiation mechanism (Figure 9). The Junonia coenia densovirus genome has an inverted terminal repeat of 517 nt, the first 96 nt of which can fold to form a T-shaped structure of the type found in the ITR of AAV DNA.
Species demarcation criteria in the genus
Members of each species are probably antigenically distinct, and natural infection is confined to a single host species. Species are <95% related by non-structural gene DNA sequence.
List of species in the genus Densovirus
| Galleria mellonella densovirus |
|
|
| Galleria mellonella densovirus | [L32896] | (GmDNV) |
| Junonia coenia densovirus |
|
|
| Junonia coenia densovirus | [S17265] | (JcDNV) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List other related viruses which may be members of the genus Densovirus but have not been approved as species
| Diatraea saccharalis densovirus | [AF036333] | (DsDNV) |
| Helicoverpa armigera densovirus |
| (HaDNV) |
| Mythimna loreyi densovirus | [AY461507] | (MlDNV) |
| Pseudoplusia includens densovirus |
| (PiDNV) |
Genus Pefudensovirus
Type species Periplaneta fuliginosa densovirus
Distinguishing features
The genome is about 5.5 kb in size, with ambisense organization (Figure 2) and relatively small ITRs (125–175 nt). The VP gene is split into a large and a small ORF (upstream) on the 5’ half of the complementary strand. Unlike other members of the family Parvoviridae that have proteins with a PLA2 domain, the pefudensoviruses have a PLA2 motif that is located in the C-terminal portion (centred 60–70 amino acid residues from the C-terminus) of the protein predicted to be translated from the small VP ORF.
Genome organization and replication
The genes for the three non-structural proteins of pefudensoviruses are organized in the same way as those in members of the genus Densovirus, and are of similar sizes. The ORFs of the split VP gene are spliced in order to code for the largest VPs. Unlike all other members of the subfamilies Parvovirinae and Densovirinae, VP1 and VP2 have different N-termini. The N-terminus of VP2 lies within the VP-A region, whereas VP1 is encoded from transcript 4 from both the VP-A and VP-B regions. The smaller VP-B region also gives rise to some NS proteins (transcripts 5 and 6). The PLA2 motif in this region is localized in the intron of the VP-B region and is therefore only present in the transcripts that use the downstream splicing sites (connecting the 2 VP ORFs) (Figure 10). Several other densoviruses have a genome organization that is identical to that of PfDNV, but their sequence identity is very low. This may be due to the variation in insect orders to which the hosts of these viruses belong.
Species demarcation criteria in the genus
Members of each species are probably antigenically distinct, and natural infection is confined to a single host species. Species are <40% related by non-structural gene DNA sequence. VP1 and VP2 have different N-termini, i.e. VP2 is not fully contained within VP1 as for all other parvovirus genera.
List of species in the genus Pefudensovirus
| Periplaneta fuliginosa densovirus |
|
|
| Periplaneta fuliginosa densovirus | [AF192260] | (PfDNV) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Pefudensovirus but have not been approved as species
| Acheta domesticus densovirus | [AX344110] | (AdDNV) |
| Blattella germanica densovirus | [AY189948] | (BgDNV) |
| Planococcus citri densovirus | [AY032882] | (PcDNV) |
List of other related viruses which may be members of the subfamily Densovirinae but have not yet been approved as a genus or a species
Among densoviruses infecting invertebrates, but not belonging to accepted genera, four groups can be discerned: the monosense Penaeus monodon DNV (PmDNV) and related viruses, Dysaphis plantaginea densovirus (DplDNV; ambisense) and related viruses, Culex pipiens densovirus and related viruses (ambisense), and a diverse group with no clear taxonomical structure. These virus groups lack significant sequence identity or genome organizational similarities among each other or with accepted genera.
The genome of PmDNV and related viruses is about 6.3 kb in size with terminal hairpins, but no ITRs. ORFs for the structural and nonstructural proteins are on the same strand. These densoviruses are at least as different in sequence from other members of the subfamily Densovirinae as they are from any member of the subfamily Parvovirinae. Populations of virions encapsidate mostly strands of negative polarity. The genome does not contain ITRs nor any sequence recognizable as a PLA2 domain. There are probably two mRNA promoters at map units 5 and 50, and the virus probably codes for a single 54 kDa capsid protein that lacks the PLA2 motif. The left and right hairpins of the genome are 136 and 170 nt long, respectively. The original name of these viruses was hepatopancreatic parvovirus (“HPV”), and a new genus has been proposed for this group.
| Penaeus chinensis densovirus | [AY008257] | (PchDNV) |
| Penaeus merguiensis densovirus | [DQ458781] | (PmeDNV) |
| Penaeus monodon densovirus | [DQ002873] | (PmoDNV) |
Dysaphis plantaginea densovirus induces a low reproduction rate in rosy apple aphids but can produce a wing morph of the host to promote dispersal. Although the sequences for DplDNV and Myzus persicae densovirus (MpDNV) are still incomplete (about 65% sequence identity), a unique expression strategy is emerging. Like for the genus Pefudensovirus members, the VP coding sequence is split into two ORFs. However, the NS1 and NS2 ORFs are split as well, and spliced to yield in-frame coding sequences. A new genus has been proposed for this group.
| Dysaphis plantaginea densovirus | [EU851411] | (DplDNV) |
| Myzus persicae densovirus | [AY148187] | (MpDNV) |
The third group contains so far a single virus, Culex pipiens densovirus (CpDNV), with a 6 kb genome. It shares some characteristics with lepidopteran viruses belonging to the genus Densovirus, such as a single ORF for VP. However, it deviates with respect to the structure of ITRs, which are significantly shorter (285 nt), lack the GAC repeats assumed to function as an NS1-binding site, and the 68 terminal nt fold back to generate a J-shaped structure. The second difference concerns the split NS1 and NS2 coding sequences each in two ORFs, implying a splicing of the mRNA to put their N-terminal and C-terminal sequences in frame. A third significant difference lies in the mode of expression of NS genes, which requires two specific promoters, one (P7) to drive expression of NS3 and one (P17) to drive expression of NS1 and NS2. A new genus has been proposed for this group.
| Culex pipiens densovirus | [FJ810126] | (CpDNV) |
Several densoviruses remain unassigned due to the lack of sequence information. Agraulis vanillae densovirus (AvDNV), Euxoa auxilliaris densovirus (EaDNV), Lymantria dispar densovirus (LdiDNV) and Pieris rapae densovirus (PrDNV) have lepidopteran hosts, whereas Leucorrhinia dubia densovirus (LduDNV) infects Odonata. LdiDNV was isolated from cell lines.
| Agraulis vanillae densovirus |
| (AvDNV) |
| Euxoa auxilliaris densovirus |
| (EaDNV) |
| Leucorrhinia dubia densovirus |
| (LduDNV) |
| Lymantria dispar densovirus |
| (LdiDNV) |
| Pieris rapae densovirus |
| (PrDNV) |
Phylogenetic relationships within the family Parvoviridae
Only the NS1 gene product has conserved motifs among all parvoviruses. Nevertheless, many features are conserved among all parvo- and densoviruses. Figure 11 shows that a low sequence identity still can yield very similar structures. For instance, the VPs of PPV and MVM can be superimposed. Despite low sequence identity (52%), 97% of the Cαs of residue pairs have similar positions (root-mean-square errors of 1.0 Å). It can be expected that proteins with sequence identities higher than 30% have quite similar structures. Even between PPV and GmDNV, 40% of their VPs residues align despite only 9% sequence identity (RMS error of 3.0 Å). Nevertheless, phylogenetic analysis (here the Phylip neighbour-joining method, Figure 11) is useful, in conjunction with genomic organizations, to distinguish the subfamiles, genera and possible new genera within the family.
Similarity with other taxa
Previously, some Bombyx mori densoviruses (BmDNV2 and BmDNV3 isolated from the silkworm) were also classified among the parvoviruses. Although they also contained a linear, ssDNA genome, this is as far as the similarity goes. Their genome is split (hence their description as bidensoviruses), totals 14 kb instead of the 4–6 kb of the parvoviruses, and has different terminal structures. Moreover, they contain a polymerase motif, in contrast to parvoviruses, no PLA2 motif and have a different capsid composition. The conserved NS1 of parvoviruses, having similar helicase and rolling-circle replication motifs, does not have a bidensovirus counterpart.
Derivation of names
Adeno: from Greek aden, “gland”.
Amdo: from Aleutian mink disease.
Bocavirus: from bovine and canine
Brevi: from Latin brevis, “short”.
Denso: from Latin densus, “thick, compact”.
Dependo: from Latin dependeo, “to hang down”.
Erythro: from Greek erythros, “red”.
Parvo: from Latin parvus, “small”.
Pefu: from Periplaneta fuliginosa densovirus.
Further reading
Brown, K.E. (2006). The genus erythrovirus. In: J.R. Kerr, S.F. Cotmore, M.E. Bloom, R.M. Linden and C.R. Parrish (Eds.), Parvoviruses. Hodder Arnold, London. pp. 25-45.
Cotmore, S.F. and Tattersall, P. (2007). Parvoviral host range and cell entry mechanisms. Adv. Virus Res., 70, 183-232.
Flegel, T.W. (2006). Shrimp parvoviruses. In: J.R. Kerr, S.F. Cotmore, M.E. Bloom, R.M. Linden and C.R. Parrish (Eds.), Parvoviruses. Hodder Arnold, London, pp. 487-493.
Gao, G., Vandenberghe, L.H., Alvira, M.R., Lu, Y., Calcedo, R., Zhou, X. and Wilson, J.M. (2004). Clades of Adeno-associated viruses are widely disseminated in human tissues. J. Virol., 78, 6381-6388.
Kapoor, A., Simmonds, P., Slikas, E., Li, L., Bodhidatta, L.,Sethabutr, O., Triki, H., Bahri O., Oderinde, B.S., vBaba, M.M., Bukbuk, D.N., Besser, J., Bartkus, J. and Delwart, E. (2010). Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J. Infect. Dis., 201, 1633-1643.
López-Bueno, A., Segovia, J.C., Bueren, J.A., O'Sullivan, M.G., Wang, F., Tattersall, P. and Almendral, J.M. (2008). Evolution to pathogenicity of the parvovirus minute virus of mice in immunodeficient mice involves genetic heterogeneity at the capsid domain that determines tropism. J. Virol., 82, 1195-1203.
Qiu, J. and Pintel, D. (2008). Processing of adeno-associated virus RNA. Front Biosci., 13, 3101-3115.
Tattersall, P. (2006). The evolution of parvovirus taxonomy. In: J.R. Kerr, S.F. Cotmore, M.E. Bloom, R.M. Linden, C.R. Parrish (Eds.), Parvoviruses. Hodder Arnold, London. pp. 5-14.
Tijssen, P., Bando, H., Li, Y., Jousset, F.-X., Zadori, F.G., El-Far, M.Z., Szelei, J. and Bergoin, M. (2006). Evolution of densoviruses. In: Kerr, J.R., Cotmore, S.F., Bloom M.E., Linden, R.M. and Parrish, C.R. (Eds.), Parvoviruses. Hodder Arnold, London, pp. 55-68.
Zadori, Z., Szelei, J., Lacoste, M.C., Li, Y., Gariepy, S., Raymond, P., Allaire, M., Nabi, I.R. and Tijssen, P. (2001). A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell, 1, 291-302.
Contributed by
Tijssen, P., Agbandje-McKenna, M., Almendral, J.M., Bergoin, M., Flegel, T.W., Hedman, K., Kleinschmidt, J., Li, Y., Pintel, D.J. and Tattersall, P.
Figures
Figure 1 Morphology of parvoviruses. (A) Side-view, at a resolution of 3.5 , of a tilted model of a porcine parvovirus (PPV) capsid with the top five trimers translated 120 along its 5-fold axis from the middle body of 10 trimers (five shown in dark blue and five in light blue); the bottom five trimers are not shown. The 5-fold axes are located at the intersection of five trimers (e.g. at arrow: green, magenta, two dark blue and one light blue colored trimer). (B) Top view of model shown in (A), without light-blue trimers, along the 5-fold axis. The channels at the 5-fold axes are clearly visible (arrow indicates same 5-fold axis as shown by arrow in (A)). (C) Structure of a PPV trimer. The arrows indicate the intertwining of the GH-loop (between strands G and H) in the counter-clockwise located proteins from near the 3-fold axis towards the 2-fold axis (arrows). The GH-loop actually consists of two loops of which one (loop 3, yellow), running to the 2-fold axis, is partly covered by loop 4 (cyan) near the 3-fold axis (oval arrow). (D) Parvoviruses may have remarkably similar structures despite low sequence identities. This figure shows the alignment of minute virus of mice (MVM, in red; 549 amino acid residues, PDB 1mvm) and PPV (in blue; 542 amino acid residues, PDB 1k3v) structural proteins that have only 52% sequence identity. Nevertheless, 528 Cs (97%) of the residue pairs occupy the same position in the capsid (root-mean-square error of 1.0 ). (E) Negative contrast electron micrograph of empty and full PPV particles (bar=50 nm) and space-filling models of the capsid structures of PPV, Galleria mellonella densovirus (GmDNV; PDB: 1DNV), human B19 virus (B19V; PDB: 1s58), and adeno-associated virus - 2 (AAV-2; PDB: 1lp3) shown at a resolution of 4 . In each case, the view is down a 2-fold axis at the centre of the virus, with 3-fold axes left and right of centre, and 5-fold axes above and below. Models (A-D) have been rendered by PyMOL and the space-filling models by CHIMERA (Multiscale Models).

Figure 2 Gene organization for members of the monosense subfamily Parvovirinae, the genera Iteravirus and Brevidensovirus, and some unassigned parvoviruses (top). Gene organization for the ambisense densoviruses and some unassigned densoviruses (bottom).
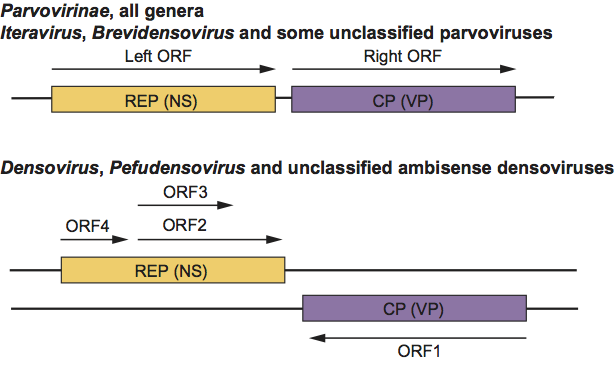
Figure 3 DNA replication model as determined for minute virus of mice (MVM). The newly synthesized DNA of the growing 3 end is represented by a black arrowed line, with the original genome in dark grey. Light grey denotes progeny genomes embedded within the oligomeric intermediates. Upper and lower cases of R and L represent flip and flop forms of the right and left ends, respectively. For details see the text.

Figure 4 Gene organization and transcription scheme for members of the genus Parvovirus, as shown for minute virus of mice (MVM). Genes are shown as boxes. The left ends of the mRNAs (thick lines) are the sites of the mRNA caps (filled circles), the right ends are the polyadenylation sites (arrows); introns are indicated by thin lines.

Figure 5 Gene organization and transcription scheme for members of the genus Erythrovirus, as shown for human parvovirus B19 (B19V). Genes are shown as boxes. The left ends of the mRNAs (thick lines) are the sites of the mRNA caps (filled circles), the right ends are the polyadenylation sites (arrows); introns are indicated by thin lines.

Figure 6 Gene organization and transcription scheme for members of the genus Dependovirus, as shown for adeno-associated virus 2 (AAV-2) and the nonprimate adeno-associated virus 2 (AAV-5). Genes are shown as boxes. The left ends of the mRNAs (thick lines) are the sites of the mRNA caps (filled circles), the right ends are the polyadenylation sites (arrows); introns are indicated by thin lines. In contrast to the primate AAVs, nonprimate AAVs also have a polyadenylation site in the middle of the genome. Both types of AAVs have 3 promoters.

Figure 7 Gene organization and transcription scheme for the member of the genus Amdovirus, as shown for Aleutian mink disease virus (AMDV). Genes are shown as boxes. The left ends of the mRNAs (thick lines) are the sites of the mRNA caps (filled circles), the right ends are the polyadenylation sites (arrows); introns are indicated by thin lines (1=R1; 2=R1; 3=R2; 4=R2; 5=Rx; 6=Rx).

Figure 8 Gene organization and transcription scheme for members of the genus Bocavirus, as shown for bovine parvovirus (BPV). Genes are shown as boxes. The left ends of the mRNAs (thick lines) are the sites of the mRNA caps (filled circles), the right ends are the polyadenylation sites (arrows); introns are indicated by thin lines

Figure 9 Gene organization and transcription scheme for members of the ambisense genus Densovirus, as shown for Galleria mellonella densovirus (GmDNV). Genes are shown as boxes as well as the ORFs that they contain. The starts of the mRNAs (thick lines) are the sites of the mRNA caps (filled circles), the ends are the polyadenylation sites (arrows); the intron is indicated by thin lines. ORF1 and ORF2/3 are translated by a leaky scanning mechanism from transcript 3 and 2, respectively.
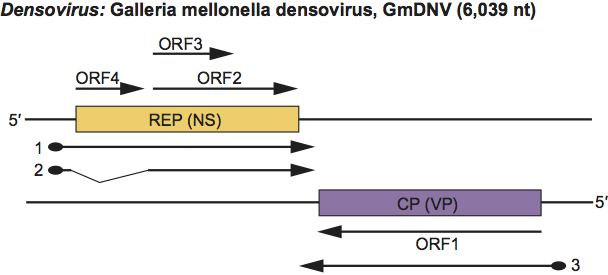
Figure 10 Gene organization and transcription scheme for members of the ambisense genus Pefudensovirus, as shown for Acheta domesticus densovirus (AdDNV). Genes are shown as boxes as well as the ORFs that they contain. The starts of the mRNAs (thick lines) are the sites of the mRNA caps (filled circles), the ends are the polyadenylation sites (arrows); the intron is indicated by thin lines. ORF1a/b and ORF2/3 are translated by a leaky scanning mechanism from transcript 4 and 2, respectively. Transcript 1 yields NS3 (ORF4), transcript 2 yields NS1 and NS2, transcript 3 yields VP2, transcript 4 yields VP1, VP3 and VP4 (possibly VP5), transcripts 5 and 6 yield also nonstructural proteins (ORF1b). There are some minor variations in the expression strategies of these viruses (e.g. an extra intron for Periplaneta fuliginosa densovirus (PfDNV) indicated by dashed lines).

Figure 11 Phylogenetic relationship among the pleiotropic NS1 proteins of members of the family Parvoviridae. The tree was constructed with the programs included in the Phylip package at the http://mobyle.pasteur.fr/cgi-bin/portal.py website (ClustalW-multialign, Phylip distance matrix, PROTDIST, Neighbor-Joining method, and phylogenetic tree drawing). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The scale bar represents the rate of amino acid substitutions. This phylogenetic analysis distinguishes the two subfamilies, as well as the monosense and ambisense densoviruses, and recognizes the different genera as separate clades. Several other clades that are possible new genera, as described in the text, are also recognized. The asterisk indicates clades of viruses that do not contain the phospholipase A2 motif in their capsid proteins.

