Family: Nanoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Distinguishing features
The family Nanoviridae comprises plant viruses possessing very small virions containing a multipartite (6–8), circular, single stranded DNA genome and being transmitted by aphids in a circulative (non-propagative) persistent manner. Each virion contains one component of the multipartite genome and 60 subunits of a capsid protein (CP) of about 19 kDa.
Virion properties
Morphology
Virions are 17–20 nm in diameter, and presumably of an icosahedral T=1 symmetry structure containing 60 subunits. They are not enveloped. Capsomeres may be evident, producing an angular or hexagonal outline (Figure 1).
Physicochemical and physical properties
Virions are stable in Cs2SO4 but may not be stable in CsCl. The buoyant density of virions is about 1.24 to 1.30 g cm−3 in Cs2SO4, and 1.34 g cm−3 in CsCl. They sediment as a single component in sucrose rate-zonal and Cs2SO4 isopycnic density gradients. The sedimentation coefficient of banana bunchy top virus (BBTV) virions is 46S. The particle weight of subterranean clover stunt virus (SCSV) is approximately 1.6×106. The extinction coefficient of SCSV is 3.6 at A260 (corrected for light scattering).
Nucleic acid
Six or eight circular single stranded (ss)DNAs ranging in size from 923 to 1111 nucleotides (nt) are consistently found in virion preparations of different babu- and nanovirus isolates. They are encapsidated as individual positive sense strands in separate particles. In addition, up to four different satellite-like ssDNAs of about 1000–1100 nt, also referred to as alphasatellites, are found associated with the majority of the nanovirid isolates.
Proteins
Virions have a single CP of about 19 kDa. No other proteins have been found associated with virions. In addition, at least 5–7 non-structural proteins are encoded by the mRNA(s) transcribed from the nanovirid ssDNAs (Table 1). All but one of the nanovirid DNAs encode only a single protein. A second virion-sense ORF, completely nested within the M-Rep-encoding ORF and encoding a putative 5 kDa protein of unknown function (U5), was identified only from banana bunchy top virus (BBTV) DNA-R but not from any other nano- and babuvirus DNA-R.
Table 1 Designation, size and functions of the proteins encoded by the various DNA components of the members of the family Nanoviridae
| Proteina | Protein size (in kDa) | Encoded by DNA component | Identified fromb | Protein function(s)c | |||
| Nanoviruses | Babuviruses | ||||||
| FBNYV | SCSV | BBTV | ABTV | ||||
| M-Rep | 33.1–33.7 | DNA-R | + | + | + | + | Replication initiator protein for all genomic DNAs |
| CP | 18.7–19.6 | DNA-S | + | + | + | + | Structural protein, virion formation (encapsidation) |
| Clink | 18.5–19.9 | DNA-C | + | + | + | + | Cell-cycle regulation |
| MP | 12.7–13.7 | DNA-M | + | + | + | + | Cell-to-cell movement |
| NSP | 17.3–17.7 | DNA-N | + | + | + | + | Presumed NSP (by analogy to geminiviruses) |
| U1 | 16.9–18.0 | DNA-U1 | + | + | − | − | Unknown |
| U2 | 14.2–15.4 | DNA-U2 | + | − | − | − | Unknown |
| U3 | 10.3 | DNA-U3 | − | − | + | − | Unknown |
| U4 | 10 or 12.5 | DNA-U4 | + | − | − | − | Unknown |
aMaster replication initiator protein (M-Rep), coat protein (CP), cell-cycle link protein (Clink), movement protein (MP) and nuclear shuttle protein (NSP). U1 to U4 are temporary designations until the protein function is determined.
b+ or − indicates whether a protein has been described from a virus species or not. Note that the genome organization of all other nanoviruses is identical to that of FBNYV.
cThe italicized letters indicate how the DNA component designation has been derived.
Lipids
Not known.
Carbohydrates
Not known.
Genome organization and replication
Based on a range of geographical isolates, there is compelling evidence that 6 and 8 DNAs are essential and form the genome of a babu- and nanovirus, respectively (Figures 2 and 3). All nanovirid DNAs have a similar structural organization, containing conserved inverted repeat sequences potentially forming a stem-loop structure that is part of the common region-stem loop [CR-SL], and a second common region named CR-M (for babuviruses) or CR-II (for nanoviruses) (Figures 2 and 3). The additional satellite-like DNAs contain different inverted repeat sequences (stem loops).
Babu- and nanoviruses share a set of five homologous DNA components, referred to as DNA-R, -S, -C, -M and -N (Figures 2 and 3). Three other DNAs (DNA-U1, -U2 and -U4) encoding proteins of as yet unknown functions have been identified from nanoviruses only and one further DNA (DNA-U3) only from babuviruses. It should be noted, however, that no ORF has been identified from the DNA-U3 of the two Abaca bunchy top virus (ABTV) isolates and several Asian isolates of BBTV. Whereas the difference in the number and types of DNAs between babu- and nanoviruses reflects a fundamental difference between viruses of these two genera, the apparent disparity in genomic organizations between some viruses of a genus may indicate that certain genome components have not yet been identified. This might be true for DNA-U2 and -U4 of SCSV and seems definitely to be the case for cardamom bushy dwarf virus (CBDV) DNAs other than DNA-R.
All nanovirid DNAs contain a major virion sense ORF and are transcribed unidirectionally. However, two mRNAs (encoding M-Rep and U5 protein) are transcribed from the BBTV DNA-R, whereas all the other babu- and nanovirus DNAs (incl. the other babu- and nanovirus DNA-R) seem to encode a single protein only. Each coding region is preceded by a promoter sequence with a TATA box and followed by a polyadenylation signal (Figures 2 and 3). For the nanovirus DNA-R, however, the position of the polydenylation signal between the TATA box and the ORF leads to transcription of the replication origin and synthesis of a terminally redundant mRNA that is capable of folding into extended secondary structures. This may be a way to regulate the expression of the encoded master replication initiator (M-Rep) protein.
Since the nanovirid DNAs and some of the biochemical events determined for nanovirid replication resemble those of the geminiviruses, their replication is also thought to occur in the nucleus through transcriptionally and replicationally active dsDNA intermediates by a rolling circle type of replication mechanism. Upon decapsidation of viral ssDNA, one of the first events is the synthesis of viral dsDNA with the aid of host DNA polymerase. As the virus DNAs have the ability to self-prime during dsDNA synthesis, it is likely that pre-existing primers are used for dsDNA replicative form (RF) synthesis, as has been shown for BBTV. From these dsDNA forms, host RNA polymerase then transcribes mRNAs encoding the M-Rep and other viral proteins. Viral DNA replication is initiated by the M-Rep protein. There is experimental evidence for faba bean necrotic yellows virus (FBNYV) and BBTV that M-Rep has DNA cleavage and nucleotidyl transfer activity in vitro and initiates the replication of all genomic DNAs. These biochemical reactions involve a conserved nonanucleotide sequence flanked by inverted repeat sequences that potentially form a stem-loop structure and are a part of the viral origin of replication (ori). This sequence arrangement including the loop-forming sequence containing the nonanucleotide TATTATTAC or TAGTATTAC is perfectly conserved in babuviruses and nanoviruses, respectively. The non-coding regions (NCRs) of all genomic DNAs of a given nanovirid share this highly conserved stem-loop region (CR-SL) that also encompasses short repeated sequences (iterons) that are presumed to be binding sites for the M-Rep protein, the only viral protein essential for nanovirus replication. All other replication proteins including DNA polymerases are provided by the host cell. Viral DNA replication is enhanced by the action of Clink, a nanovirid-encoded cell cycle modulator protein.
In addition to the genomic DNAs, a number of additional DNAs encoding Rep proteins have been found associated with some, but not all nanovirid infections. These additional DNAs, referred to as “alphasatellites” (see chapter on “Satellites”), are genetically very diverse and phylogenetically distinct from the DNA-R of nano- and babuviruses. Tentative evidence suggests that these DNAs interfere with the establishment and expression of nanovirid disease symptoms.
Antigenic properties
Virions are strong immunogens. Most viruses belonging to the same genus are serologically related to, but distinct from, one another. No serological relationship between babu- and nanoviruses has been observed.
Biological properties
Host range
Viruses of the individual species have narrow host ranges. Nanoviruses naturally infect only a limited range of leguminous species (Fabaceae), whereas babuviruses have been reported only from few monocots, such as the Musaceae and Zingiberaceae. All viruses are associated with stunting of infected plants, and infected hosts may also show leaf roll, chlorosis and premature death. Nanovirids are restricted to the phloem tissue of their host plants and are not transmitted mechanically or through seeds. Until recently, plants could only be experimentally infected by graft or vector transmission. However, infectivity of purified FBNYV virions by biolistic bombardment has now been demonstrated and aphid-transmissible FBNYV and faba bean necrotic stunt virus (FBNSV) virions have been reconstituted using eight cloned DNAs.
Transmission
Under natural conditions, all viruses are transmitted by certain aphid species in a persistent manner and do not replicate in their vectors. Together with results from complementation experiments, there is evidence that vector transmission of FBNYV (and probably also other nanovirids) require a virus-encoded helper factor that is either dysfunctional or absent in purified virion preparations.
Geographical distribution
While BBTV is widely distributed in banana growing countries in the Asia-Pacific region and Africa, ABTV has been reported only from Sarawak (Malaysia) and the Philippines, and CBDV only from India. SCSV occurs in Australia, milk vetch dwarf virus (MDV) in China and Japan, pea necrotic yellow dwarf virus (PNYDV) in Europe and FBNSV in Ethiopia and Morocco. In contrast, the reported geographic distribution of FBNYV is much wider, occurring in several countries of West Asia and North and East Africa as well as in a European country (Spain). No nanovirus has been recorded from the New World.
Genus and species demarcation criteria in the family
Features demarcating the two genera in the family, i.e., criteria to be used for distinguishing babuviruses from nanoviruses, are shown in Table 2.
Criteria to be used as guidelines for species demarcation are:
- Differences in natural host range
- Differences in the number and types of vector aphid species
- Different reactions to antibodies to individual species
- Differences in CP aa sequences of >15%, and/or
- Overall nt sequence identity of <75% is generally indicative of a distinct species.
Since several nanovirids are now known to have overlapping host ranges and to be transmitted by a similar range of aphid species, biological criteria appear no longer useful for species discrimination within a genus. Although species-specific monoclonal antibodies (where available) can be used for species discrimination, preference should nowadays be given to the molecular criteria specified above.
Table 2 Features distinguishing viruses of the genera Nanovirus and Babuvirus
| Features | Genus Nanovirus | Genus Babuvirus |
| Major hosts | Dicots (legumes) | Monocots |
| Major aphid vectors | Aphis craccivora and a few other legume-colonizing aphid species | Pentalonia and Micromyzus spp. |
| No. of genomic DNAs | 8 | 6 |
| Presence of specific DNAs | DNA-U1, -U2, -U4 | DNA-U3 |
| Size of genomic DNAs | 923–1020 nt | 1013–1111 nt |
| Overall nucleotide sequence relationship in DNA-R, -N, -S, -C and -M | >50% (between species) | >50% (between species) |
| <45% (between babu- and nanoviruses) | ||
Genus Nanovirus
Type species Subterranean clover stunt virus
Distinguishing features
Experimental and circumstantial evidence suggests that the nanovirus genome consists of eight different ssDNA components, referred to as DNA-R, -S, -M, -C, -N, -U1, -U2 and -U4 (Figure 2). Thus, the latter three DNAs are characteristic for viruses of the genus Nanovirus. The other major differences between babu- and nanoviruses are that the latter naturally infect legumes (dicots), are vectored by several aphid species colonizing legumes, and share low levels of aa sequence identities ranging from 18 to 56% in individual genes with babuviruses. Moreover, the nanovirus DNA components ranging in size from 923 to 1020 nt are slightly smaller (by ca. 100 nt) than those of babuviruses (1013–1111 nt) and, in contrast to babuviruses, the nanovirus DNA-R transcripts are terminally redundant.
Antisera to FBNYV and SCSV cross-react weakly with SCSV and FBNYV respectively in Western blots and immunoelectron microscopy, but not at all in DAS-ELISA, suggesting that the serological relationship between these two viruses is distant (CP aa sequence identity ca. 57%). However, MDV and FBNSV react strongly not only with FBNYV antisera but also with the majority of monoclonal antibodies to FBNYV. Therefore, species-specific MAbs are required for the differentiation and specific detection of these three closely related species, which share CP aa sequence identities of 83–85%.
Nanoviruses have largely overlapping but relatively narrow host ranges. They infect over 50 legume species and only a few non-legume species under experimental and natural conditions. They are transmitted by several aphid species. Aphis craccivora appears to be the major natural vector of these viruses as it is the most abundant aphid species on legume crops in the afflicted areas and was among the most efficient vectors under experimental conditions. Other aphid vectors are Acyrthosiphon pisum and Aphis fabae but for MDV also A. gossypii and Megoura viciae.
List of species in the genus Nanovirus
| Faba bean necrotic yellows virus |
|
|
| Faba bean necrotic yellows virus-[Egypt] | [AJ132179 to -84, AJ132186, AJ749902] | (FBNYV-[EG]) |
| Faba bean necrotic yellows virus-[Morocco] | [GQ274023 to -30] | (FBNYV-[MA]) |
| Milk vetch dwarf virus |
|
|
| Milk vetch dwarf virus-[Japan] | [AB000923 to -7; AB009046, AB027511, AB044387, AB255373] | (MDV-[JP]) |
| Subterranean clover stunt virus |
|
|
| Subterranean clover stunt virus-[Australia] | [U16730, U16732–4, U16736, AJ290434] | (SCSV-[AU]) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Nanovirus but have not been approved as species
| Faba bean necrotic stunt virus |
|
|
| Faba bean necrotic stunt virus-[Ethiopia] | [GQ150778 to -85] | (FBNSV-[ET]) |
| Faba bean necrotic stunt virus-[Morocco] | [GQ274031 to -8] | (FBNSV-[MA]) |
| Pea necrotic yellow dwarf virus |
|
|
| Pea necrotic yellow dwarf virus-[Germany] | [GU553134*] | (PNYDV-[DE]) |
* Although only a DNA-R sequence has been described for PNYDV-[DE] (Grigoras et al., Plant Dis.94, 643, 2010), the seven other genome segments have been sequenced and aphid transmissible virus has been reconstituted using cloned DNAs.
Genus Babuvirus
Type species Banana bunchy top virus
Distinguishing features
Babuviruses naturally infect monocots (Musa and Amomum spp.) and are vectored by the banana aphid Pentalonia nigronervosa (CBDV also by Micromyzus kalimpongensis Basu). BBTV and ABTV infect bananas (Musa spp.) and closely related species within the Musaceae, such as M. textilis Née and Ensete ventricosum Cheesem. There are no confirmed non-Musa hosts of BBTV and ABTV. Symptoms of BBTV include plant stunting, foliar yellowing and most characteristic dark green streaks on the pseudostem, petioles and leaves. There is not much information on CBDV, which has recently been found associated with a serious disease (“Foorkey”) of large cardamom (Amomum subulatum Roxb.) in India.
BBTV and ABTV have a genome consisting of six different ssDNA components, referred to as DNA-R, -S, -C, -M, -N, and -U3 (Figure 3). Since they have been identified from all babuvirus isolates studied in greater detail, they are considered integral components of the babuvirus genome. Although there are several BBTV isolates from which a DNA-U3 has not been identified, most babuviruses seem to possess a specific DNA component (DNA-U3) of unknown function. Only for DNA-U3 of BBTV-[AU] (L41576) has an RNA transcript been reported.
Unlike the relative conservation among the genes within nanovirus species, there is considerable variation in certain genes among individual isolates of a babuvirus species. Striking genetic differences ranging from 7 to 20% have been observed for the individual gene products of two ABTV isolates, one from the Philippines and the other from Sarawak (Malaysia). Similarly, two groups of BBTV isolates, designated the Asian and South Pacific groups, are distinguished based on DNA-R, -N and -S sequences. The nt sequences of the major gene of DNA-R, -N and -S differ by 7.5, 8.6 and 6.3% which translate to mean differences of 5.6, 6.7 and 1.4%, respectively, in aa sequences between the two geographic groups. Particularly striking are the differences between the two groups of BBTV isolates in the CR-M of DNA-R (32%), -N (27%) and -S (39%), whereas the intra-group CR-M variation does not exceed 6%. Whereas the DNA-R, -N and -S genes appear to be well conserved among BBTV isolates, striking differences (up to 19%) between BBTV isolates in less conserved gene products (e.g., movement protein) have been observed (Figure 4). If these genetic differences can be substantiated by completely sequencing a number of Asian isolates, this might form the basis for a future subdivision of BBTV into two separate species.
BBTV is serologically unrelated to members of the genus Nanovirus. However, BBTV antibodies have been used for the detection of ABTV and CBDV, the two other babuviruses. Of 10 monoclonal antibodies raised to BBTV, only two reacted with ABTV. This is consistent with a CP amino acid sequence difference of about 20% between ABTV and BBTV.
List of species in the genus Babuvirus
| Abaca bunchy top virus |
|
|
| Abaca bunchy top virus-[Malaysia] | [EF546808 to -13] | (ABTV-[MY]) |
| Abaca bunchy top virus-[Philippines] | [EF546802 to -7] | (ABTV-[PH]) |
| Banana bunchy top virus |
|
|
| Banana bunchy top virus-[Australia] | [S56276, L41574 to -78] | (BBTV-[AU]) |
| Banana bunchy top virus-[Taiwan] | [DQ826390 to -1; DQ826393 to -6] | (BBTV-[TW]) |
| Cardamom bushy dwarf virus |
|
|
| Cardamom bushy dwarf virus-[India] | [AY485960*] | (CdBDV-[IN]) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Although the DNA-R sequence available from GenBank is defective, an apparently complete DNA-R sequence has been determined.
List of other related viruses which may be members of the genus Babuvirus but have not been approved as species
None reported.
List of unassigned species in the family Nanoviridae
| Coconut foliar decay virus |
|
|
| Coconut foliar decay virus-[Vanuatu] | [M29963*] | (CFDV-[VU]) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Since this appears to be an alphasatellite sequence, it should no longer be assigned to the family Nanoviridae.
Phylogenetic relationships within the family
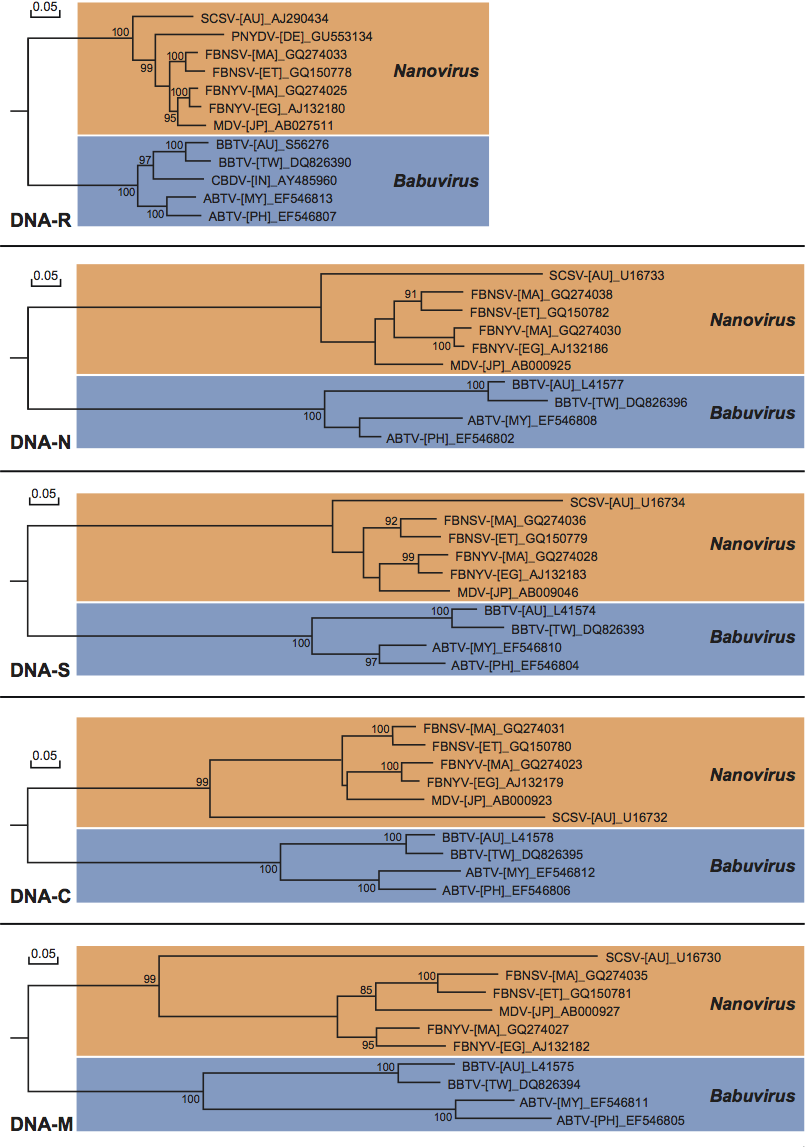
Analysis of the five proteins identified from the majority of the assigned nanovirid species (Figure 4) suggests that the most conserved nanovirid proteins are the M-Rep protein (54–97% identity) and the NSP (41–91%), followed by the CP (20–84%), the MP (14–76%) and the Clink protein (18–72%). Babuviruses share significant levels of aa sequence similarity with nanoviruses only in the M-Rep (54–56%) and NSP (41–45%), whereas the aa sequence similarities between the two genera are negligible in the CP (20–27%), the MP (20–23%) and the Clink protein (18–23%).
Similarity with other taxa
All Rep proteins of the assigned species have most of the aa domains characteristic of Rep proteins of geminiviruses and other ssDNA viruses. The nanovirus Rep proteins differ from those of members of the family Geminiviridae in being smaller (about 33 kDa), having a slightly different dNTP-binding motif (GPQ/NGGEGKT), lacking the retinoblastoma-like protein (Rb)-binding motif (LxCxE) and sharing aa sequence identities of only 17 to 22% with them. Moreover, the assigned species are clearly distinct from geminiviruses in particle morphology, genome size, number and size of DNA components, mode of transcription, and in vector species. Although circo- and nanovirids possess closely related Rep proteins and morphologically similar virions, circovirids infect vertebrates and have a much smaller genome (1.8–2.3 kb) that is not only monopartite but also bidirectionally transcribed. All these ssDNA viruses have a conserved nonanucleotide motif at the apex of the stem-loop sequence which is consistent with the operation of a rolling circle model for DNA replication.
Derivation of names
Babu: from banana bunchy top virus.
Nano: from the Greek nanos, meaning “dwarf”, referring to the observations that these plant viruses have the smallest known virions and genome segment sizes, and dwarf their hosts.
Further reading
Burns, T.M., Harding, R.M. and Dale, J.L. (1995). The genome organization of banana bunchy top virus: analysis of six ssDNA components. J. Gen. Virol., 76, 1471-1482.
Chu, P.W.G. and Vetten, H.J. (2003). Subterranean clover stunt virus. AAB Descriptions of Plant Viruses, 396.
Grigoras, I., Gronenborn, B. and Vetten, H.J. (2010). First report of a nanovirus disease of pea in Germany. Plant Dis., 94, 643.
Grigoras, I., Timchenko, T., Grande-Pérez, A., Katul, L., Vetten, H.J. and Gronenborn, B. (2010). High variability and rapid evolution of a nanovirus. J. Virol., 84, 9105-9117.
Grigoras, I., Timchenko, T. and Gronenborn, B. (2008). Transcripts encoding the nanovirus master replication initiator protein are terminally redundant. J. Gen. Virol., 89, 583-593.
Grigoras, I., Timchenko, T., Katul, L., Grande-Pérez, A., Vetten, H.J. and Gronenborn, B. (2009). Reconstitution of authentic nanovirus from multiple cloned DNAs. J. Virol., 83, 10778-10787.
Hu, J.M., Fu, H.C., Lin, C.H., Su, H.J. and Yeh, H.H. (2006). Reassortment and concerted evolution in banana bunchy top virus genomes. J. Virol., 81, 1746-1761.
Karan, M., Harding, R.M. and Dale, J.L. (1994). Evidence for two groups of banana bunchy top virus isolates. J. Gen. Virol., 75, 3541-3546.
Sharman, M., Thomas, J.E., Skabo, S. and Holton, T.A. (2008). Abacá bunchy top virus, a new member of the genus Babuvirus (family Nanoviridae). Arch. Virol., 153, 135-147.
Vetten, H.J. (2008). Nanoviruses. In: Encyclopedia of Virology (B.W.J Mahy and M.H.V. Van Regenmortel, Eds), 3rd edn. Elsevier Oxford, vol. 3, pp. 385-391.
Contributed by
Vetten, H.J., Dale, J.L., Grigoras, I., Gronenborn, B., Harding, R., Randles, J.W., Sano, Y., Thomas, J.E., Timchenko, T. and Yeh, H.H.
Figures
Figure 1 Negative contrast electron micrograph of particles of Faba bean necrotic yellows virus (FBNYV). The bar represents 50 nm.
(Courtesy of L. Katul and D.-E. Lesemann.)
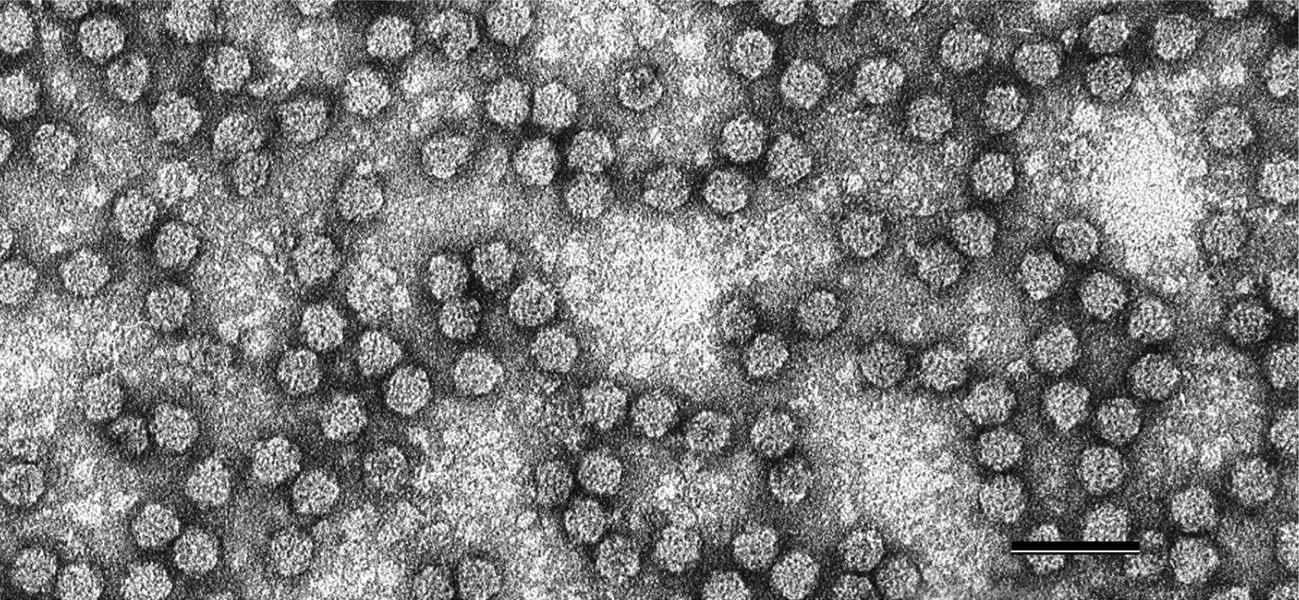
Figure 2 Diagram illustrating the genomic organization of viruses of the genus Nanovirus and depicting the structure of the eight identified viral DNA components (see also Table 1). Each DNA circle (100020 nt) contains its designated name and the name of the encoded protein. Arrows refer to the location and approximate size of the ORFs and the direction of transcription. Note that DNA-U2 and -U4 have not been identified from SCSV. The position of the common stem-loop region (CR-SL) and the second common region (CR-II) are indicated.
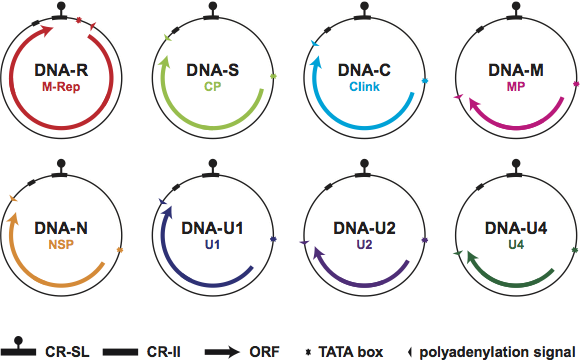
Figure 3 Diagram illustrating the genomic organization of viruses of the genus Babuvirus and depicting the structure of the six identified viral DNA components (see also Table 1). Each DNA circle (106050 nt) contains its designated name and the name of the encoded protein. The position of the common stem-loop region (CR-SL) and the major common region (CR-M) are indicated. Arrows refer to the location and approximate size of the ORFs and the direction of transcription. Note that (i) no ORF has been identified from DNA-U3 of the two ABTV isolates and several Asian isolates of BBTV and (ii) DNA-R of BBTV has a small internal ORF which potentially encodes a 5 kDa protein (U5) and has been shown to be transcribed from DNA-R of BBTV-[AU].
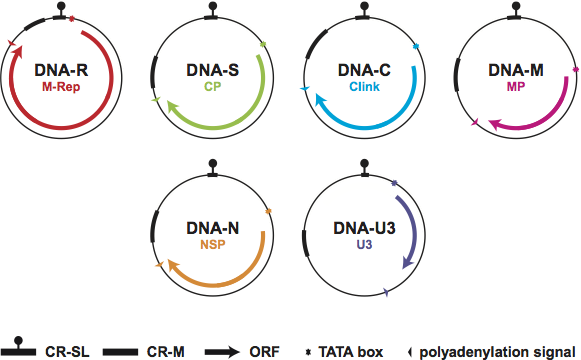
Figure 4 Neighbor-joining dendrograms illustrating nucleotide sequence relationships in the five DNAs (DNA-R, -N, -S, -C and -M) identified from all assigned and (still) putative members of the genera Nanovirus and Babuvirus (family Nanoviridae): abaca bunchy top virus (ABTV), banana bunchy top virus (BBTV), cardamom bushy dwarf virus (CBDV), faba bean necrotic yellows virus (FBNYV), faba bean necrotic stunt virus (FBNSV), milk vetch dwarf virus (MDV), pea necrotic yellow dwarf virus (PNYDV), and subterranean clover stunt virus (SCSV). Since sequence information is available for genetically distinct isolates of some nano- and babuviruses and this may have future taxonomic implications, the sequences of ABTV isolates from Malaysia (MY) and the Philippines (PH), BBTV isolates from Australia (AU) and Taiwan (TW), FBNYV isolates from Egypt (EG) and Morocco (MA), and FBNSV isolates from Ethiopia (ET) and Morocco (MA) were included in the comparison. Vertical branch lengths are arbitrary and horizontal distances are proportional to the number of base substitutions per site (see scale bar). Sequence alignments and dendrograms were produced using DNAMAN (version 6, Lynnon Biosoft, Quebec, Canada) which uses a CLUSTAL-type algorithm. The dendrograms were bootstrapped 1000 times (scores are shown at nodes).