Family: Microviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Members of the family Microviridae are non-enveloped, ssDNA, prokaryotic viruses with T=1 icosahedral symmetry. There are two morphologies represented within the family Microviridae (Figure 1), which along with genome organization, sequence homologies and host lifestyle, separate the family into two distinct parts, the genus Microvirus and the three genera of subfamily Gokushovirinae. Members of the genus Microvirus infect enterobacteria, and share a morphology and genome organization typified by Enterobacteria phage φX174 (φX174). Members of the Gokushovirinae infect obligate intracellular parasitic bacteria (Bdellovibrio and Chlamydia) and mollicutes (Spiroplasma), and share the morphology, typified by Spiroplasma phage 4 (SpV4).
Phage φX174 typifies microvirus morphology, in which pentamers of a major spike protein decorate the five-fold axes of symmetry of the T=1 lattice. The structures of φX174, Enterobacteria phage α3 (α3) and Enterobacteria phage G4 (G4) capsids (the G4 capsids were empty particles) have been determined to at least 3.5 Å resolution. CPs can be superimposed with root mean square deviations less than or equal to 0.8 Å and exhibit the common β-barrel motif. Capsids have a diameter of 250 Å. The 70 Å-diameter spike protein pentamers rise 30 Å from the surface of the capsid. Virions of the other three genera lack major spike proteins and, hence, their five-fold axes of symmetry are not decorated. Cryo-EM image reconstruction of SpV4 reveals mushroom-shaped protrusions at the three-fold axes of symmetry, which rise 54 Å above the surface of the 270 Å diameter capsids. These three-fold related structures appear to be composed of three interacting CPs and are formed by a distinct insertion loop unique to the gokushovirus CPs.
Physicochemical and physical properties
The buoyant densities of family members range from 1.38–1.41 g cm−3 for microviruses and 1.30–1.31 g cm−3 for bdellomicroviruses and chlamydiamicroviruses. The only known spiromicrovirus, SpV4, has a reported buoyant density of 1.40 g cm−3. Particles of both morphologies are very stable, resistant to detergents, ether, chloroform, pH 6.0–9.0 and freezing. Microviruses have S values of about 115S, while phages in the other genera sediment with S values near 90S.
Nucleic acid
Genomes are circular positive sense ssDNA molecules. As with morphological, biochemical and biophysical properties, genome sizes appear to fall into two size ranges. Microvirus genomes are 5.3–6.1 kb, while gokushovirus genomes are considerably smaller, 4.4–4.9 kb. The smaller genomes reflect the absence of genes encoding major spike and external scaffolding proteins.
For those genera in which multiple species have been sequenced (Microvirus and Chlamydiamicrovirus), genome arrangement is close to identical within the genus. The genome arrangement of the only sequenced member of bdellomicroviruses, Bdellovibrio phage φMH2K (φMH2K), is extremely similar to that found in chlamydiamicroviruses, with the exception of the location of gene 5.
Proteins
Although only the φX174-like phages have been studied in detail, sequence similarities and structurally based computational analyses have led to reasonable hypotheses regarding some of the viral proteins in the three less-studied gokushoviruses (Table 1).
Table 1 Proteins found in members of the family Microviridae
| Microvirus proteins | Chlamydiamicrovirus and Bdellomicrovirus proteins1 | Spiromicrovirus proteins | Protein function |
| A | Vp4 | Vp2 | DNA replication protein |
| A* |
|
| Function unknown, non-essential |
| B | Vp3 |
| Internal scaffolding protein |
| C | Vp5 |
| ssDNA synthesis, inhibitor of dsDNA synthesis |
| D | Absent | Absent | External scaffolding protein |
| E | OrfN product in φMH2K |
| Lysis protein |
| F | Vp1 | Vp1 | Major capsid protein |
| G | Absent | Absent | Major spike protein |
| H | Vp2 | Vp4 | Minor spike protein, DNA pilot protein. De novo H protein synthesis is required for efficient viral protein synthesis |
| J | Orf8 product | Orf8 product | DNA binding protein |
| K |
|
| Burst size modulation (host specific), non-essential |
| L, M | 6, 7 (Chp2) | 3,5,6,7,8,9 | Orfs of unknown coding capacity and/or proteins of unknown function |
|
| W, X, Y, Z (φMH2K) |
|
|
1 Italics indicate a hypothesized homolog. Blank entries indicate that the identity of the homolog, if it exists, is not readily apparent. Absent indicates that no homolog exists.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
Genome organization is summarized in Figure 2. DNA replication and capsid assembly has been studied only for microviruses. Considering the distant relationship with the other genera, generalizations regarding common mechanisms within the family are not appropriate. Therefore, replication is discussed at the genus level.
Antigenic properties
Both neutralizing and non-neutralizing monoclonal antibodies have been produced against microviruses. These antibodies often cross-react with other genus members. Non-neutralizing monoclonal antibodies have been produced against chlamydiamicrovirus proteins. Antibodies against the Chp2 CP recognize the φMH2K CP, further demonstrating the close relationship between these two genera.
Genus Microvirus
Type species Enterobacteriophage phiX174
Distinguishing features
All current members of the genus Microvirus were isolated from Enterobacteriaceae. However, it should be noted that rigorous searches for members of the family Microviridae have not been conducted in other hosts. Since most isolation procedures are optimized for large dsDNA viruses, which are considerably denser than microviruses, easier to visualize by electron microscopy, and have much larger S values, special techniques must be employed for the isolation of members of the family Microviridae.
Although the viruses in this group are dependent on the same host cell proteins for replication, genetic studies suggest that individual phages may be particularly sensitive to host cell alleles of rep, slyD and mraY. Even though the primary sequences of the microvirus proteins have diverged, some of these proteins can cross-function (e.g., DNA binding proteins and internal scaffolding proteins), indicating productive use; while other proteins cross-inhibit (e.g., external scaffolding proteins), indicating the ability to interact with other proteins across species lines. However, after interaction, other functions required for productive morphogenesis are hindered. Cross-species inhibitory protein domains have been identified with the use of chimeric proteins.
Genome organization and replication
Microvirus genome organization is strictly conserved in the 47 known genome sequences. The genomes most distinguishing feature is the presence of overlapping reading frames. Seven of the 11 genes (genes A through E and gene K) reside in such regions (Figure 2). Most of these genes code for non-structural proteins (Table 1). Genes found within the coding sequences of other genes (A*, B, K and E) encode non-essential proteins (A* and K), proteins that do not affect particle formation, such as lysis proteins (E); or highly flexible proteins that tolerate substitutions, like the internal scaffolding protein (B). Moreover, multiple mutant strains that no longer require B protein function have been isolated. Packaged microvirus genomes do not form the densely condensed cores seen in most dsDNA bacteriophages. Instead, genomes are intimately associated with the capsid’s inner surface, which contributes to late stages of morphogenesis and virion stability. This association, as opposed to fixed capsid dimensions, may hinder the acquisition of new genes (or morons), but has not appeared to hinder horizontal gene exchanges between different microvirus species.
Procapsid morphogenesis and DNA synthesis proceed independently of each other during microvirus replication. After phage ssDNA enters the cell, stage I DNA replication commences. This process converts the infecting (+) single stranded circular genome into a covalently closed double stranded molecule called replicative form I DNA (RFI). No viral proteins are involved. With the synthesis of the (−) strand, transcription can begin. Although microvirus gene expression is not dependent on elaborate trans-acting mechanisms to ensure temporal regulation, the relative timing and amounts of viral proteins synthesized is controlled by highly sophisticated sets of cis-acting promoters, transcription terminators and ribosome binding sites. Stage II DNA synthesis is dependent on the viral A protein, which cleaves the RFI DNA (+) strand at the origin of replication and covalently attaches itself to the DNA. This generates RF II molecules. De novo (+) strand replication involves a rolling circle mechanism, while de novo (−) strand synthesis occurs via a mechanism similar to stage I replication. Stage II DNA synthesis continues until one copy of viral protein C binds displaced ssDNA at the initiation of another round of stage II DNA synthesis, which terminates further dsDNA replication. With the binding of the C protein, the stage III DNA pre-initiation complex forms, consisting of one copy each of protein A, protein C and the host cell rep protein, a DNA helicase which associates with protein A during stage II DNA synthesis.
The packaging mechanisms involved in members of the family Microviridae differ substantially from those found in other bacteriophages. The pre-initiation complex associates with the viral procapsid (see below), forming the 50S complex in which ssDNA is concurrently synthesized and packaged. Genome length is strictly governed by a single origin of replication, which determines both the initiation and termination of biosynthesis and packaging. DNA concatamers, unique translocating vertices, and head-full mechanisms are not involved. The viral A protein bound to the origin of replication in the RF II DNA is both necessary and sufficient for packaging specificity. Although any circular DNA molecule with a microvirus origin of replication can serve as a template, the secondary structure formed by the packaged genome can affect the biophysical properties of the resulting virion. After one round of rolling circle synthesis, protein A cuts the newly generated origin and acts as a ligase, generating a covalently closed circular molecule.
The first assembly intermediates in procapsid morphogenesis are 9S and 6S particles, pentamers of viral coat and spike proteins, respectively (Figure 3). Five internal scaffolding proteins (protein B) bind to the underside of a 9S particle, which produces the 9S* particle. The upper surface of the viral coat protein can now interact with spike and external scaffolding proteins. 9S* particles are also less prone to aggregate prematurely than 9S particles and the presence of protein B may facilitate the incorporation of the minor vertex protein, or DNA pilot protein, protein H. Twelve 12S* particles then associate with external scaffolding proteins (protein D) to form the procapsids. In the φX174 procapsid crystal structure, four D proteins, as dimers of dimers, are associated with one CP. After particle formation, two-fold related B-B and D-D scaffolding contacts keep the CP pentamers from dissociating during DNA packaging.
The genome replication/packaging machinery binds to a depression along the two-fold axis of symmetry and an ssDNA genome is then concurrently synthesized and packaged, along with 60 copies of the DNA binding protein, protein J. Procapsids are probably filled through one of the 30 Å diameter pores at the three-fold axes of symmetry. During packaging, B proteins are extruded, probably displaced by the DNA binding protein J, which shares a binding cleft in the viral CP. Finally, D proteins dissociate, yielding the virion. This is accompanied by an 8.5 Å radial collapse of CPs around the packaged genome, which is tethered to the underside of the CP pentamers by F-DNA and J-DNA interactions. Genetic and biochemical data indicate that capsid-ssDNA interactions may mediate the integrity of this final stage of morphogenesis.
Biological properties
The nature of the interaction of φX174-like phages with their hosts is poorly understood. Initial attachment to host cells occurs via a sugar residue, most likely glucose, in the lipopolysaccharide (LPS). A site on the surface of the capsid, near the three-fold axes of symmetry, has been shown to bind glucose reversibly. However, host range mutations change amino acids in spike proteins G and H, suggesting that a second host cell receptor may be required for DNA ejection. The identity of this second factor is unknown. Although the members of the family Microviridae are tailless, the virus may follow a pathway similar to that of the large-tailed Enterobacteria phage T4 (T4). T4 reversibly interacts with LPS via its long tail fibers. The phage then “walks” along the surface of the cell until it finds a second receptor, which triggers ejection. Instead of walking, microviruses may “rock and roll” along the cell surface, until this second receptor is found.
Intracellular localization of φX174-like phage maturation is strongly dependent on the host cell rep allele, which must physically interact with the viral A protein and the procapsid during ssDNA synthesis and packaging. Proper interactions between the viral E protein and the gene products of host cell slyD and mraY alleles are probably required for lysis.
Species demarcation criteria in the genus
Currently, species demarcation criteria are temperature and host range. However, both phenotypes can be changed by single point mutations. Therefore, these criteria may not be rigorous for distinguishing between species. The results of phylogenetic analyses of 42 novel isolates suggest that the microviruses fall into three clades represented by bacteriophage φX174, α3 and G4. Thus, it may be more appropriate to consider most isolates as varieties of these three phages.
List of species in the genus Microvirus
| Enterobacteria phage alpha3 |
|
|
| Enterobacteria phage α3 | [X60322] | (α3) |
| Enterobacteria phage G4 |
|
|
| Enterobacteria phage G4 | [J02454] | (G4) |
| Enterobacteria phage phiX174 |
|
|
| Enterobacteria phage φX174 | [J02482] | (φX174) |
| Enterobacteria phage S13 | [M14428] | (S13) |
| Enterobacteria phage phiK |
|
|
| Enterobacteria phage φK | [X60323] | (φK) |
| Enterobacteria phage St-1 |
|
|
| Enterobacteria phage St-1 | [GQ149088] | (St-1) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Microvirus but which have not yet been approved as species
None reported.
Subfamily Gokushovirinae
Taxonomic structure of the subfamily
Subfamily Gokushovirinae
Genus Chlamydiamicrovirus
Genus Bdellomicrovirus
Genus Spiromicrovirus
Distinguishing features
The members of the subfamily Gokushovirinae differ from those in the genus Microvirus in virion morphology (see Figure 1), genome organization (see Figure 2), sequence homologies and host lifestyle. They infect obligate intracellular parasitic bacteria and mollicutes whereas microviruses infect enterobacteria. The three genera are distinguished by host organism.
Genus Chlamydiamicrovirus
Type species Chlamydia phage 1
Distinguishing features
These phages infect various species of Chlamydia and Chlamydophila. Stocks of the type species phage no longer exist. Computational analyses indicate that chlamydiamicrovirus capsids will resemble those of SpV4.
Genome organization and replication
The genome organization of the Chlamydiamicrovirus Chp-2 is depicted in Figure 2. Since double stranded replicative form DNA has been isolated and homologs of microvirus proteins A and C are present, DNA replication is thought to occur via a similar mechanism. The mechanisms involved in capsid formation in the genus Chlamydiamicrovirus are not known, but will probably not resemble microviruses morphogenesis because chlamydiamicroviruses lack external scaffolding and major spike proteins. However, Vp3 has been demonstrated to be an internal scaffold protein. The Chp-2 viral lifecycle has been characterized and is tightly regulated with the developmental cycle of its host.
Biological properties
Unlike the microviruses, the chlamydiamicroviruses probably recognize a protein receptor.
Species demarcation criteria in the genus
There are no formal criteria for species demarcation.
List of species in the genus Chlamydiamicrovirus
| Chlamydia phage 1 |
|
|
| Chlamydia phage 1 | [D00624] | (Chp-1) |
| Chlamydia phage 2 |
|
|
| Chlamydia phage 2 | [AJ270057] | (Chp-2) |
| Chlamydia pneumoniae phage CPAR39 |
|
|
| Chlamydia pneumoniae phage CPAR39 | [AE002163] | (φCPAR39) |
| Guinea pig Chlamydia phage |
|
|
| Guinea pig Chlamydia phage | [U41758] | (φCPG1) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Chlamydiamicrovirus but which have not yet been approved as species
None reported.
Genus Bdellomicrovirus
Type species Bdellovibrio phage MAC 1
Distinguishing features
These phages infect Bdellovibrio strains. DNA of isolates of the type species, Bdellovibrio phage MAC 1, has not been sequenced and stocks of this phage no longer exist. All bdellovibriovirus genome and biophysical data come from the study of φMH2K. Computational analyses suggest that the structure of φMH2K resembles SpV4.
Genome organization and replication
The genome organization of the φMH2K is depicted in Figure 2. Replication is most likely similar to Chp-2.
Species demarcation criteria in the genus
There are no formal criteria for species demarcation.
List of species in the genus Bdellomicrovirus
| Bdellovibrio phage phiMH2K |
|
|
| Bdellovibrio phage φMH2K | [AF306496] | (φMH2K) |
| Bdellovibrio phage MAC 1 |
|
|
| Bdellovibrio phage MAC 1 |
| (MAC-1) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Bdellomicrovirus but which have not yet been approved as species
None reported.
Genus Spiromicrovirus
Type species Spiroplasma phage 4
Distinguishing features
The one isolated phage of this group infects Spiroplasma melliferum.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Spiromicrovirus
| Spiroplasma phage 4 |
|
|
| Spiroplasma phage 4 | [M17988] | (SpV4) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Spiromicrovirus but which have not yet been approved as species
None reported.
Phylogenetic relationships within the family (Figure 4)
Phylogenetic trees built with four different proteins (CP, Rep protein, DNA pilot protein and the internal scaffolding protein) clearly illustrate the existence of the two major groups of the family.
Similarity with other taxa
In some instances ssDNA genomes of viruses in the family Microviridae are similar in organization to those of members of the family Inoviridae. Structurally, viruses in the family Microviridae resemble those in the family Parvoviridae. Atomic structures of the CPs are rich in insertion loops coming off the β-barrel core. Similarities between the capsids of spiromicroviruses, bdellomicroviruses, chlamydiamicroviruses and parvoviruses are more pronounced due to the complex interactions of CPs at the three-fold axes of symmetry.
Derivation of names
Micro: from the Greek micros, “small”.
Gokusho: from the Japanese Gokusho, “very small” or “very holy”.
Further reading
Brentlinger, K., Hafenstein, S., Novak, C.R., Fane, B.A., Birgon, R., McKenna, R. and Agbandje-McKenna, M. (2002). Microviridae, a family divided. Isolation, characterization and genome sequence of a φMH2K, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovorus. J. Bacteriol., 184, 1089-1094.
Chen, M., Uchiyama, A. and Fane, B.A. (2007). Eliminating the requirement of an essential gene product in an already very small virus: scaffolding protein B-free φX174, B-free. J. Mol. Biol., 373, 308-314.
Cherwa, J.E., Jr., Sanchez-Soria, P., Wichman, H.A. and Fane, B.A. (2009). Viral adaptation to an antiviral protein enhances the fitness level to above that of the uninhibited wild type. J. Virol., 83, 11746-11750.
Chipman, P.R., Agbandje-McKenna, M., Renaudin, J., Baker, T.S. and McKenna, R. (1998). Structural analysis of the Spiroplasma virus, SpV4, implications for evolutionary variation to obtain host diversity among the Microviridae. Structure, 6, 135-145.
Dokland, T., McKenna, R., Ilag, L.L., Bowman, B.R., Incardona, N.L., Fane, B.A. and Rossmann, M.G. (1997). Structure of a viral procapsid with molecular scaffolding. Nature, 389, 308-313.
Fane, B.A., Brentlinger, K.L., Burch, A D., Hafenstein, S L., Moore, E., Novak, C.R. and Uchiyama, A. (2006). φX174 et al. In: The Bacteriophages. (R. Calendar, ed). Oxford Press, London, pp. 129-145.
Hafenstein, S. and Fane, B.A. (2002). φX174 genome-capsid interactions influence the biophysical properties of the virion: evidence for a scaffolding-like function for the genome during the final stages of morphogenesis. J. Virol., 76, 5350-5356.
Rokyta, D.R., Burch, C.L., Caudle, S B. and Wichman, H.A. (2006). Horizontal gene transfer and the evolution of microvirid coliphage genomes. J. Bacteriol., 188, 1134-1142.
Salim, O., Skilton, R.J., Lambden, P.R., Fane, B.A. and Clarke, I.N. (2008). Behind the chlamydial cloak: the replication cycle of chlamydiaphage Chp2, revealed. Virology, 377, 440-445.
Zheng, Y., Struck, D.K., Bernhardt, T.G. and Young, R. (2008). Genetic analysis of MraY inhibition by the phiX174 protein E. Genetics, 180, 1459-1466.
Contributed by
Cherwa, J.E. Jr. and Fane, B.A.
Figures
Figure 1 (Top left) Cryo-image reconstructions of the two morphologies represented within the family Microviridae: (left) Spiroplasma phage 4 (SpV4) (genus Spiromicrovirus); (right) Enterobacteria phage X174 (X174) (genus Microvirus) (courtesy T. Baker, R. McKenna and M.G. Rossmann). (Top right) Negative contrast electron micrograph of X174 particles. The bar represents 50 nm. (Bottom) Electronic rendering surface of X174 particles (left, scaffold; center, procapsid) and diagram representing the T=1 lattice.
(Dokland et al. (1997). Nature, 389, 308313)
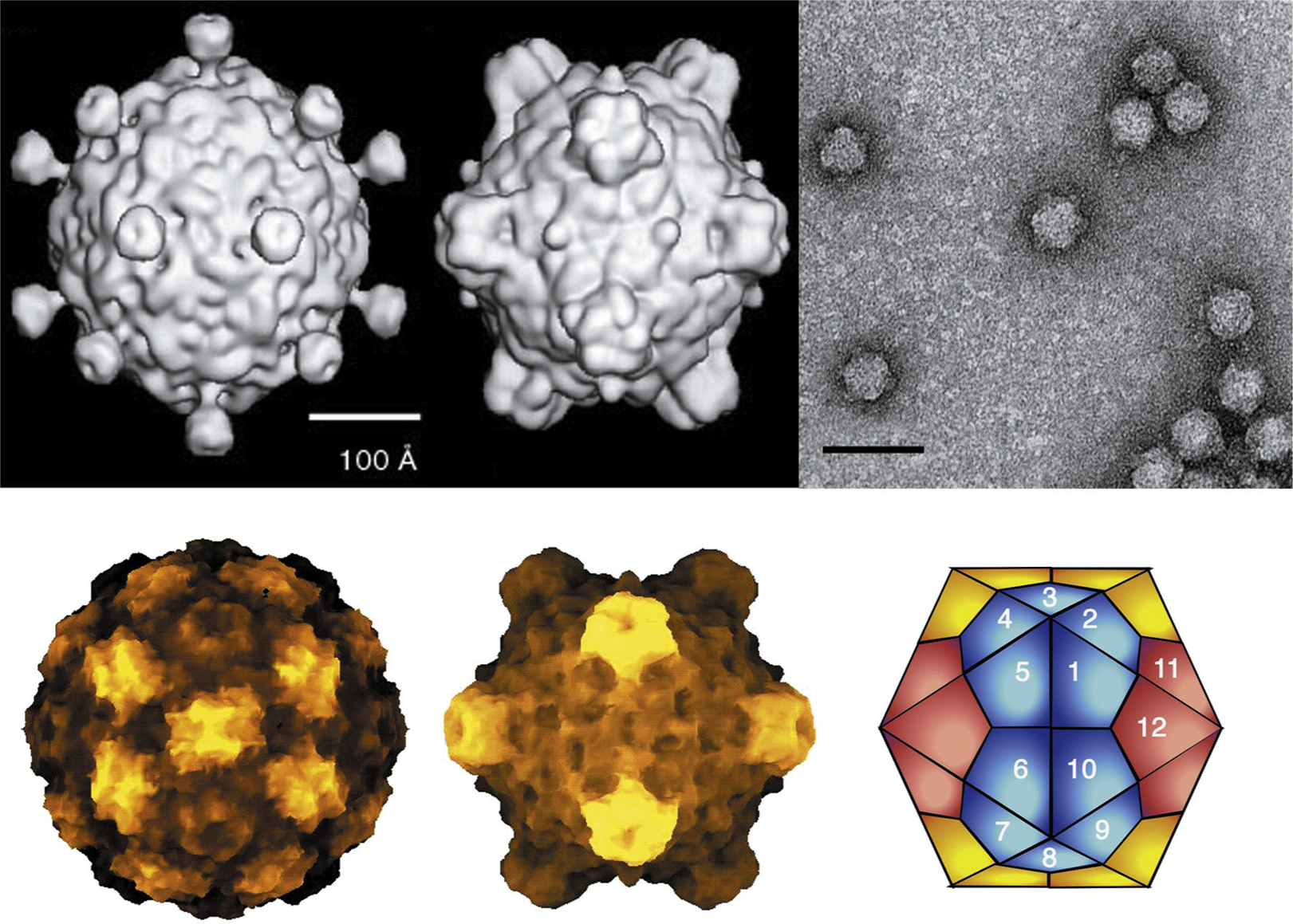
Figure 2 Genome organization of members of the family Microviridae. The circular genomes are presented linearly. Due to pronounced protein homologies and genome arrangements, the same gene number scheme is used for both the bdellomicroviruses (Bdellovibrio phage MH2K; MH2K) and chlamydiamicroviruses (Chlamydia phage 2; Chp2). Four proteins in Spiroplasma phage 4 (SpV4), (the products of genes 1, 2, 4, and 8) have homologs in the chlamydia- and bdellomicroviruses, the number in parentheses indicates the homologous gene.
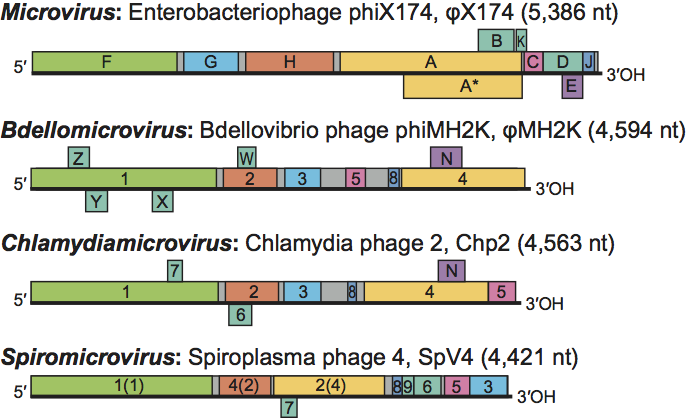
Figure 3 Microvirus capsid morphogenesis.
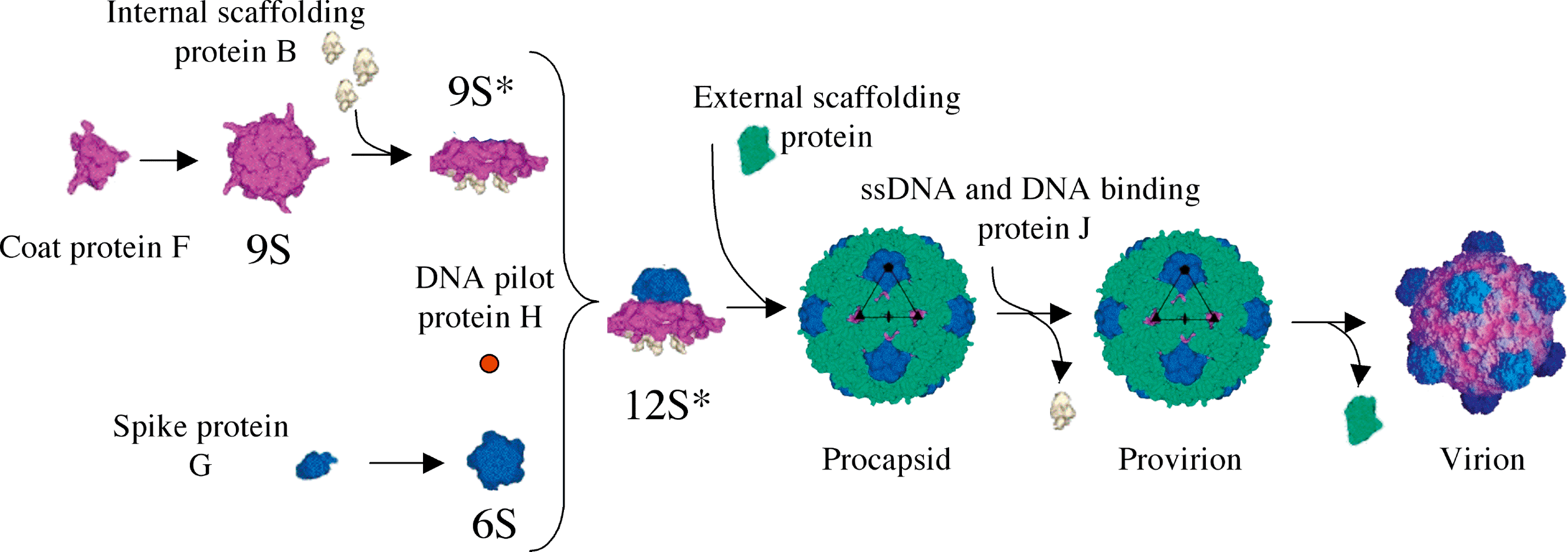
Figure 4 (opposite) Phylogeny of the family Microviridae. The aa sequences of the capsid protein (tTop left), the Rep protein (top right), the DNA pilot protein (bottom left) and the internal scaffolding protein (bottom right) were used to make alignments with the CLUSTAL X software. The trees were designed with PAUP and the bootstrap values are indicated above 50%. The abbreviations of the viruses used and their GenBank accession numbers are listed in the List of Species of the description.