Family: Inoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
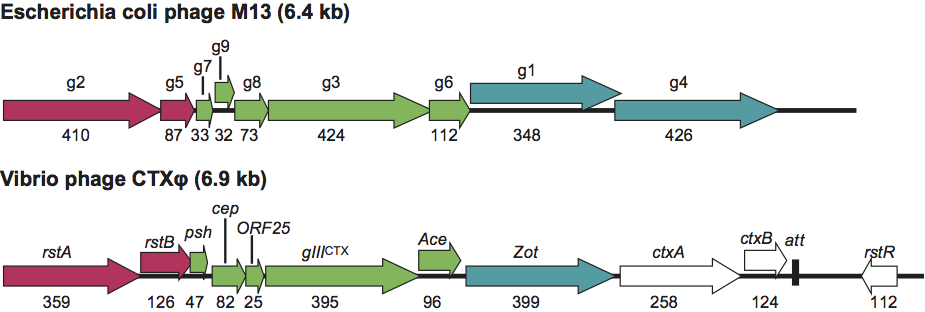
Virions in this family contain a circular, positive sense, single stranded DNA genome within a helical array of thousands of copies of a major capsid protein. Inoviruses are flexible filaments about 7 nm in diameter that infect gram-negative and gram-positive bacteria, while plectroviruses are short rods about 15 nm in diameter that infect mycoplasmas (Figure 1). The packaged loops of DNA have anti-parallel strands extending between the virion ends, with fold-backs at the ends. The fold-back at one end is a specific DNA sequence that initiates packaging, and that at the other end is a random sequence that triggers the addition of a multi-domain adsorption protein. A crystal structure has been determined for the adsorption protein of one inovirus, but otherwise little is known about the structures of end proteins. In micrographs, the initiating ends of inoviruses are blunt, while the adsorption ends are tapered with extensions, as shown in Figure 1b. The virions are extruded, as shown in Figure 1c for plectrovirus progeny emerging from innumerable sites on the membrane of the wall-less mycoplasma host. Inoviruses of gram-negative hosts assemble and extrude from a more limited number of sites at adhesions between outer and inner membranes.
Virion contour lengths depend on DNA size and on the average nucleotide rise, h, characterizing the DNA conformation. Lengths of inoviruses range from 700 nm for Pseudomonas phage Pf3 (5833 nt, h=0.24 nm), to 900 nm for E. coli phages in the Ff group (f1, fd, and M13; 6407 nt, h=0.28 nm) and up to 3700 nm projected for Pseudomonas phage Pf4 (12437 nt, h=0.61 nm). Major capsid subunits (gp8 or equivalent) have from 42 to 57 amino acids, the N-terminal domains of which maintain virion solubility, while hydrophobic central domains stabilize subunit–subunit interactions, and C-terminal domains interact with the DNA. The gene 8 subunits are highly α-helical. They overlap each other in arrays in either Class I symmetry or Class II symmetry. Class I capsids have subunits arranged in steps of two interdigitated pentamers, and Class II capsids have subunits arranged in helices of approximately 5.4 interdigitated monomers per turn. The DNA helices within many of these capsids have bases stacked at the center and H-bonded, albeit with only about 25% Watson–Crick H-bonding possible, but otherwise similar to classical right-handed A- and B-form DNA helices. The DNA helix in Escherichia coli phage fd has a nucleotide rotation of 36° and a rise of 0.28 nm. However, the Pseudomonas phage Pf1 has an everted (inside-out), highly twisted and stretched DNA helix with a nucleotide rotation of 131.8° and a rise of 0.61 nm; the sugar-phosphate backbones are in the center and the bases are on the outside. Coat protein sequence similarities indicate Pf1-like DNA conformations in Psuedomonas phage Pf4 and Vibrio phage f237 (VfO3K6). Atomic models of inovirus structures are available in the structure databases, and refined models based on X-ray and spectroscopic date are continually being developed.
Plectrovirus virions are nearly straight rods with one end rounded and the other more variable. Acholeplasma phages are 70–90 nm long and 15 nm in diameter, whereas Spiroplasma phages are 230–280 nm long and 10–15 nm in diameter (Figure 1, lower). Negative stained images suggest 4±2 nm hollow cores. Optical diffraction of images of Acholeplasma phage MV-L1 suggest morphological units arranged with two-fold rotational and 5.6-fold screw symmetry. DNA conformations in this genus have apparent nucleotide rise values, h, near 0.07 nm.
Physicochemical and physical properties
Members of the family Inoviridae are sensitive to chloroform and ether, but have differing sensitivities to detergents. They are resistant to wide ranges of pH, and to extremes of cold and heat. The inovirus Pf1 has a unique thermally induced virion symmetry transition near 10 °C that is of considerable value to structural studies. Inoviruses have DNA contents from 6% to 14% and buoyant densities in CsCl in the narrow range 1.28±0.02 g cm−3. Virions range in total mass from 10 MDa to over 60 MDa, but S20,w values are in a narrow range 40–45S because mass-per-length values are in a narrow range near 18,000 Da nm−1. Translational and rotational diffusion constants are consistent with persistence lengths on the order of their contour lengths. Plectrovirus buoyant densities are 1.39 g cm−3 in CsCl and 1.21 g cm−3 in metrizamide, as reported for Spiroplasma phage SpV1-KC3. Virions in the family are easily precipitated from dilute solutions by low concentrations of polyethylene glycol.
In addition to random flexing, most inoviruses exhibit large-length-scale coiling. Strong coiling is evident for E. coli phages X and C-2 in electron micrographs, and gentle coiling is evident in liquid crystal behaviour for phage fd and several others. However, Pseudomonas phage Pf1 shows no evidence of the phenomenon. The varying degrees of coiling depend on the varying characteristics of capsid–DNA interactions, including the stoichiometric ratio of nucleotides per subunit, the value of which lies between 2 and 2.5 for many species, but which is uniquely 1 (unity) for Pf1.
Nucleic acid
The genomes in the family range in size from 4.5 kb to as much as 12.4 kb and encode from 4 to 17 or more genes. Virions, as well as assembly precursor complexes in the cytoplasm, contain one molecule of positive sense ssDNA. In some cases genes are expressed from complementary strands, in particular the repressor genes when the viral genome is a latent prophage integrated in the cellular genome.
Proteins
In the genus Inovirus, the type virus M13 has five different proteins in its virion (Figures 1 and 2). The cylindrical shells are composed of 2700 copies of gp8 (5.2 kDa), the adsorption end has probably five each of gp3 (43 kDa) and gp6 (12 kDa), and the assembly nucleation end has probably five each of gp7 (3.5 kDa) and gp9 (3.3 kDa).
In the genus Plectrovirus, the Spiroplasma phages SpV1-R8A2B and SpV1-T78 have major capsid proteins of 7.5 kDa, while the Acholeplasma phages MVL1 and MVL51 have major coat proteins apparently of 19 kDa, according to gel data, although there is a strong tendency to aggregate.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
Inovirus genomes all have core regions similar to the genome of type virus M13 which is comprised of a DNA replication module (g2, g5 and g10), a capsid protein module (g7, g9, g8, g3 and g6), a morphogenesis module (g1,g4 and g11), and intergenic module with control sequences (Figure 2). Genes are closely spaced, even overlapping. Many inoviruses are integrative phages present in the host chromosome as latent prophages with their core regions flanked by genes for integrases, repressors, transposases and virulence factors such as toxin genes, as well as additional control sequences, such as operators and insertion sequences. An example is the well-characterized genome of vibriophage CTX, in which the genes for the subunits of cholera toxin are in the locus of a key morphogenesis gene of M13. Extensive sequence homologies between non-integrative and integrative phages, as between Pseudomonas phages Pf1 and Pf4, and many other pairs, indicate close evolutionary relationships. Members of the family Inoviridae do not lyse their hosts, so the established terms “lytic” and “lysogenic” are not fully suitable, but they are commonly used and not likely to be misunderstood.
Non-integrative, productive infections invariably involve the following steps: (1) phage adsorption to specific cell surface receptors, usually plasmid encoded conjugative pili, then binding to periplasmic protein co-receptors that couple adsorption with DNA uptake into the cytoplasm; (2) host enzyme conversion of the released ssDNA to an initial dsDNA circle from which viral proteins are expressed; (3) semi-conservative DNA replication initiated by viral endonuclease at a specific origin sequence, concomitant with continued expression of structural and assembly proteins; (4) rolling circle synthesis of progeny ssDNA that become sequestered by viral ssDNA binding protein; (5) and finally membrane-based assembly and extrusion of virions without cell lysis. Once infection is established virus extrusion continues indefinitely as cells divide. Primary control of expression is transcriptional from promoters of various strengths around the genome. Translational controls use overlapping reading frames and alternate starts in the same frame. Intergenic regions contain the complementary-strand and viral-strand replication origins and DNA packaging signals. Cell growth rates are usually slowed enough on lawns to generate plaques, but phage production on lawns without plaque formation can occur.
Amongst members of the family Inoviridae, the integration of genomes into host chromosomes was first reported for Xanthomonas phages Cf1t and Cf16 and for the SpV1-type Spiroplasma phages. The Cf1 insertions were at one bacterial site, whereas the plectrovirus insertions were at 17 or more sites in their plant pathogenic mycoplasmas. These insertions are mediated by viral integrases and transposases. In contrast, insertions mediated by bacterial host XerCD recombinases were later observed for vibriophage/prophage CTXφ, the pathogenicity island of V. cholera carrying the cholera toxin genes, and for prophages CUS1 and CUS2, which are virtually identical elements bearing an apparent virulence factor in the genomes of the human pathogens E. coli O18:K1:H7 and Y. pestis. Thus, the mechanisms for conversions into prophage states, as well as mechanisms of maintenance and induction vary considerably within the family Inoviridae. Vibriophage CTX infections have been the most extensively studied. They start with virus binding to TCP pili receptors followed by ssDNA release and formation of dsDNA, as described above for non-integrative infections. Holliday junctions can then form between att sites on phage dsDNA and the bacterial dif sites. Repeated formations of junctions produce tandem, or multiple, insertions which allow the replication of circular viral DNA from the linear chromosome site. Maintenance of the prophage state is through the combined actions of a phage-encoded repressor, RstR, and a host-encoded repressor, LexA. Induction of the prophage state to phage and toxin production occurs when the pathogenic bacterium colonizes the gut.
Known plectroviruses infect members of the genera Acholeplasma and Spiroplasma. Their surface receptors are not well characterized, but may have both polysaccharide as well as protein components. The replication pathways of Acholeplasma phage MV-L51 and the SpV1 group of Spiroplasma phages are similar to each other and to inoviruses with respect to freely replicating circular dsDNA and to the assembly and release of progeny virions at the membrane while the cells continue to divide. Genomes of SpV1-type phages contain insertion sequences and transposase genes, as do chromosomes of mycoplasmas infected by them. Certain high passage mycoplasma strains contain the latent prophages without producing virus, and these strains produce turbid plaques when super-infected with isolated virus. Mechanisms of insertion, repression, immunity and induction amongst the systems used by members of the family Inoviridae present many open questions.
Biological properties
Members of the family Inoviridae mobilize DNA in the microbial world, and thus play a role in the evolution of microorganisms. They do not lyse their hosts, so that progeny cells remain infected, whether in a virulent productive state or in a latent prophage state. Thus vertical transfers of genomes continually occur, and horizontal transfers occur through the extruded virions. Members of the family are found in most, if not all, ecological niches of the planet, being present in all manner of commensal and pathogenic species in plants and animals, and surviving free under a range of environmental conditions. Most are either themselves integrative phages, or they are phylogenetically related to integrative phages. They carry virulence factors and are implicated in the pathogenesis of plant diseases including wilts and cankers, and of animal diseases including cholera, cystic fibrosis, acute gastroenteritis, neonatal meningitis, gonorrhea and plague. In many cases, the pathogenicity depends on the induction of a latent prophage.
The structures and lifestyles of these phages make them particularly well suited for gene transfer functions. Their virions can lengthen to carry extra genes and can adapt adsorption specificities to new hosts. In some cases their genomes can be mobilized in alternate capsids, and the physicochemical nature of DNA packaging can evolve to adapt to new environments. With regard to the latter, the types of DNA-protein interaction stabilizing virions vary from predominantly electrostatic, as in phages of the Ff group and many others, to predominantly hydrophobic, as in KSF-1, which has no basic amino acid residues in its major capsid protein. The properties are exemplified in several systems, such as by the presence of Pseudomonas prophage Pf4 (12.4 kb) in the genome of pathogenic Ps. aeruginosa strain O1 (PAO1), carrying 5.1 kb of control functions and virulence factors, on a virtually unmodified core genome of Pseudomonas phage Pf1 (7.3 kb) specific for Ps. aeruginosa strain K (PAK). Further, the existence of virtually identical prophages CUS1 and CUS2 in the genomes of the pathogens E. coli O18:K1:H7 and Y. pestis, mentioned above, points to significant roles for inoviruses in pathogen evolution. Contributing to these roles are cellular receptors that are conjugative type IV pili encoded by transmissible plasmids so that lateral transfer of these plasmids concomitantly transfers phage sensitivity to new hosts. Of special importance in regard to host range among bacteria of the genus Vibrio, is that vibriophages fs1, KSF-1, VEJ, VGJ, and VSK all use as receptors the mannose-sensitive hemagglutinin (MSHA) pili present on many vibrio strains, and all of them are xer-recombinase intergrative phages capable of transducing genetic elements. All of them contribute to the diversity of the Vibrionacea. And finally, as a plectrovirus example, many pathogenic Spiroplasma citri strains from plants bear freely replicating circular DNA and extrude SpV1-like virions while also bearing prophages in the chromosomes of continually dividing mycoplasma cells (Figure 1).
Genus Inovirus
Type species Enterobacteria phage M13
Distinguishing features
Virions are slender and flexible 7±1 nm filaments having a range of lengths, but narrow range of mass-per-length values near 18,000 Da nm−1, hence a narrow range of sedimentation coefficients near 42 S20,w. Nucleic acid contents are low, 6–14% by weight, keeping buoyant densities in CsCl in the range of 1.28±0.02 g cm−3. Hosts are gram-negative and gram-positive bacteria.
Species demarcation criteria within the genus
- Host range
- Non-exchangeability of structural genes
Species demarcations are not definitive because of mosaicism generated by widespread lateral gene transfers and host chromosome integration as described above. Inovirus species are listed under the bacterial host species for the initial isolate together with incompatibility group plasmid that defines host ranges.
List of species in the genus Inovirus
| 1. Phages ofEnterobacteriaceae |
|
|
|
| Enterobacteria phage AE2 | {IncF} |
|
|
| Enterobacteria phage AE2 |
|
| (AE2) |
| Enterobacteria phage C-2 | {IncC} |
|
|
| Escherichia coli phage C-2 |
|
| (C-2) |
| Enterobacteria phage dA | {IncF} |
|
|
| Enterobacteria phage dA |
|
| (dA) |
| Enterobacteria phage Ec9 | {IncF} |
|
|
| Enterobacteria phage Ec9 |
|
| (Ec9) |
| Enterobacteria phage f1 | {IncF} |
|
|
| Escherichia coli phage f1* |
| [J02448] | (f1) |
| Enterobacteria phage fd | {IncF} |
|
|
| Escherichia coli phage fd* |
| [J02451] | (fd) |
| Enterobacteria phage HR | {IncF} |
|
|
| Enterobacteria phage HR |
|
| (HR) |
| Enterobacteria phage I2-2 | {IncI2} |
|
|
| Escherichia coli phage I2-2 |
| [X14336] | (I2-2) |
| Enterobacteria phage If1 | {IncI} |
|
|
| Escherichia coli phage If1 |
| [ U02303] | (If1) |
| Enterobacteria phage IKe | {IncI2, IncN, IncP-1} |
|
|
| Escherichia coli phage IKe |
| [X02139] | (IKe) |
| Enterobacteria phage M13 | {IncF} |
|
|
| Escherichia coli phage M13* |
| [V00604] | (M13) |
| Enterobacteria phage PR64FS | {IncI} |
|
|
| Enterobacteria phage PR64FS |
|
| (PR64FS) |
| Enterobacteria phage SF | {IncS} |
|
|
| Enterobacteria phage SF |
|
| (SF) |
| Enterobacteria phage tf-1 | {IncT} |
|
|
| Escherichia coli phage tf-1 |
|
| (tf-1) |
| Enterobacteria phage X | {IncX, IncI2, IncN, IncP-1} |
|
|
| Escherichia coli phage X |
|
| (X) |
| Enterobacteria phage X-2 | {IncX} |
|
|
| Escherichia coli phage X-2 |
|
| (X-2) |
| Enterobacteria phage ZJ/2 | {IncF} |
|
|
| Escherichia coli phage ZJ/2 |
|
| (ZJ/2) |
| 2. Phages ofPseudomonadaceae |
|
|
|
| Pseudomonas phage Pf1 | {PAK} |
|
|
| Pseudomonas phage Pf1 |
| [X52107] | (Pf1) |
| Pseudomonas phage Pf2 |
|
|
|
| Pseudomonas phage Pf2 |
|
| (Pf2) |
| Pseudomonas phage Pf3 | {PA01} |
|
|
| Pseudomonas phage Pf3 |
| [M11912] | (Pf3) |
| 3. Phages ofVibrionaceae |
|
|
|
| Vibrio phage 493 |
|
|
|
| V. cholera phage/prophage 493 |
|
| (493) |
| Vibrio phage CTX |
|
|
|
| V. cholera phage/prophage CTXφ |
| [GU942563] | (CTX) |
| Vibrio phage fs1 |
|
|
|
| V. cholera phage/prophage fs1 |
| [D89074] | (fs1) |
| Vibrio phage fs2 |
|
|
|
| V. cholera phage fs2 |
| [AB002632] | (fs2) |
| Vibrio phage v6 |
|
|
|
| Vibrio phage v6 |
|
| (v6) |
| Vibrio phage Vf12 |
|
|
|
| V. parahaemolyticus phage Vf12 |
| [AB012574] | (Vf12) |
| Vibrio phage Vf33 |
|
|
|
| V. parahaemolyticus phage Vf33 |
| [AB012573] | (Vf33) |
| Vibrio phage VSK |
|
|
|
| V. cholera phage VSK |
| [AF453500] | (VSK) |
| 4. Phages ofXanthomonadaceae |
|
|
|
| Xanthomonas phage Cf16 |
|
|
|
| Xanthomonas phage/prophage Cf16 |
|
| (Cf16) |
| Xanthomonas phage Cf1c |
|
|
|
| Xanthomonas phage/prophage Cf1c |
| [M57538, U41819] | (Cf1c) |
| Xanthomonas phage Cf1t |
|
|
|
| Xanthomonas phage/prophage Cf1t |
| [U08370] | (Cf1t) |
| Xanthomonas phage Cf1tv |
|
|
|
| Xanthomonas phage/prophage Cf1tv |
|
| (Cf1tv) |
| Xanthomonas phage Lf |
|
|
|
| Xanthomonas phage/prophage Lf |
| [U10884, U38235, X70327-31, AF018286] | (Lf) |
| Xanthomonas phage Xf |
|
|
|
| Xanthomonas phage Xf |
|
| (Xf) |
| Xanthomonas phage Xfo |
|
|
|
| Xanthomonas phage Xfo |
|
| (Xfo) |
| Xanthomonas phage Xfv |
|
|
|
| Xanthomonas phage Xfv |
|
| (Xfv) |
Species names are in italic script; names of isolates are in roman script. Sequence accession n umbers [ ] and assigned abbreviations ( ) are also listed. For the many phages of Enterobacteriaceae (Group 1) the transmissible plasmids that largely define host ranges are listed in the second column.*Phages f1, fd and M13 have virtually identical structures and their 6.4 kb genomes differ at only about 1% of the nucleotide positions. They are often referred to, collectively or individually, as Ff phage. It is expected that they will be grouped into a single species in a future taxonomic revision.
List of other related viruses which may be members of the genus Inovirus but which have not yet been approved as species
| Phages of Enterobacteriaceae |
|
|
| E. coli O18:K1:H7 phage/prophage CUS1 |
| (CUS1) |
| Phages of Neissericeae |
|
|
| N. menigitidis prophage MDA | [NC_003116] | (MDA) |
| N. menigitidis prophage Nf1-A | [NC_003116] | (Nf1A) |
| N. menigitidis prophage Nf3-A | [NC_003116] | (Nf3A) |
| N. menigitidis prophages Nf1-B1,B2 | [NC_003112] | (Nf1B) |
| N. menigitidis prophages Nf1-C1,C2,C3,C4 | [NC_008767] | (Nf1C) |
| N. gonorrhea prophages Nf4-G2,G3,G5 | [NC_002946] | (Nf4G) |
| Phages of Pseudomonadaceae |
|
|
| Ps. aeruginosa phage/prophage Pf4 | [AE004091] | Pf4 |
| Ps. aeruginosa phage/prophage Pf5 | [NC_008463] | Pf5 |
| Ps. aeruginosa phage/prophage Pf7 | [CP000744] | Pf7 |
| Ps. aeruginosa prophage Pf-LESB58 | [FM209186] | (Pf-LES) |
| Phages of Ralstonaceae |
|
|
| R. solanacearum phage/prophage RSM1 | [AB259123] | (RSM1) |
| R. solanacearum phage/prophage RSS1 | [AB259124] | (RSS1) |
| R. pickettii phage/prophage p12J | [AY374414] | (p12J) |
| Phages of Shewanellaceae |
|
|
| S. piezotolerans phage/prophage SW1 | [CP000472] | (SW1) |
| Phages of Stenotrophomonaceae |
|
|
| S. maltophilia phage SMA9 | [AM040673] | (SMA9) |
| Phages of Thermusacea |
|
|
| T. thermophilus phage PH75 |
| (PH75) |
| Phages of Vibrionaceae |
|
|
| V. parahaemolyticus phage VfO4K68 | [AB043679] | (VfO4K68) |
| V. parahaemolyticus phage/prophage VfO3K6 (f237) | [AB043678] [BA000031] | (VfO3K6) (f237) |
| V. cholera phage/prophage KSF-1 | [AY714348] | (KSF-1) |
| V. cholera phage/prophage VEJ | [FJ904927] | (VEJ) |
| V. cholera phage/prophage VGJ | [AY242528] | (VGJ) |
| V. cholera phage VSKK | [AF452449] | (VSKK) |
| Phages of Xyllelaceae |
|
|
| X. fastidiosa phage Xff1 |
| (Xff1) |
| Phages of Yersiniaceae |
|
|
| Y. pestis phage/prophage CUS2 | [NC_003143] | (CUS2) (Yp01) (Ypf) |
| Phages of Gram positive bacteria |
|
|
| Clostridium beijerinickii phage CAK1 |
| (CAK1) |
| Propionibacterium freudenreichii phage B5 | [AF428260] | (B5) |
Genus Plectrovirus
Type species Acholeplasma phage MV-L51
Distinguishing features
Virions are rods with lengths of about 300 nm or less and diameters of about 15 nm. Virions are resistant to non-ionic detergents (Nonidet P-40 and Triton X-100) and sensitive to chloroform and ether. Adsorption is to cell membranes of wall-less mycoplasma hosts. Buoyant densities are 1.39 g cm−3 in CsCl and 1.21 g cm−3 in metrizamide, as reported for Spiroplasma phage SpV1-KC3.
Species demarcation criteria in the genus
- Host range
Acholeplasma viruses infect some Acholeplasma laidlawii strains. SpV1-like viruses have broad host ranges among Spiroplasma species S. citri, S. melliferum and S. kunkelii.
List of species in the genus Plectrovirus
| 1. Phages ofAcholeplasmaspp. |
|
|
| Acholeplasma phage MV-L51 |
|
|
| Acholeplasma phage MV-L51 |
| (MVL51) |
| Acholeplasma phage/prophage MV-L1 | [NC_001341] | (MVL1) |
| 2. Phages ofSpiroplasmaspp. |
|
|
| Spiroplasma phage 1-aa |
|
|
| Spiroplasma phage SpV1-aa |
| (SpV1-aa) |
| Spiroplasma phage 1-C74 |
|
|
| Spiroplasma phage/prophage SpV1-C74 | [U28974] | (SpV1-C74) |
| Spiroplasma phage 1-KC3 |
|
|
| Spiroplasma phage/prophage SpV1-KC3 |
| (SpV1-KC3) |
| Spiroplasma phage 1-R8A2B |
|
|
| Spiroplasma phage/prophage SpV1-R8A2B | [X51344] | (SpV1-R8A2B) |
| Spiroplasma phage 1-S102 |
|
|
| Spiroplasma phage/prophage SpV1-S102 |
| (SpV1-S102) |
| Spiroplasma phage 1-T78 |
|
|
| Spiroplasma phage/prophage SpV1-T78 |
| (SpV1-T78) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations () are also listed.
List of other related viruses which may be members of the genus Plectrovirus but which have not yet been approved as species
| Spiroplasma phage SkV1-CR2-3x | [EF506570] | (SkV1-S2) |
| Spiroplasma phage/prophage SVGII3 | [AJ969242] | (SVGII3) |
| Spiroplasma phage/prophage SVTS2 | [AF133242] | (SVTS2) |
List of unassigned species in the family Inoviridae
None reported.
Phylogenetic relationships within the family
Phylogenetic distances between KSF-1 and nine other filamentous vibriophages showed distinctly different cluster patterns depending upon which of three genes were used in the analysis, namely any of a gene for replication, one for phage morphogenesis, or one for receptor binding. The results showed the evolutionary effects of horizontal gene transfers resulting in mosaicism amongst the vibriophages. Some phages currently classified as different species are very closely related in all their genes (e.g. E. coli phages f1, fd and M13) and should be grouped into a single species in a future taxonomic revision.
Similarity with other taxa
The repressed latent prophage states of many members of the family Inoviridae are similar to those of temperate phages in other taxa in being inducible to virulent states that produce copius progeny, but they are distinctly different in not causing cell lysis. Hence the terms like “lysogenic” and “lytic” states are not strictly appropriate despite their wide usage.
Derivation of names
Ino: from Greek nos, “muscle filament”.
Plectro: from Greek plektron, “small stick”.
Further reading
Bille, E., Zahar, J.-R., Perrin, A., Morelle, S., Kriz, P., Jolley, K.A., Maiden, M.C.J., Dervin, C., Nassif, X. and Tinsley, C.R. (2005). A chromosomally integrated bacteriophage in invasive meningococci. J. Exp. Med., 201, 1905-1913.
Campos, J., Martnez, E., Izquierdo, Y. and Fando, R. (2010). VEJφ, a novel filamentous phage of Vibrio cholerae able to transduce the cholera toxin genes. Microbiology, 156, 108-115.
Day, L.A., Marzec, C.J., Reisberg, S.A. and Casadevall, A. (1988). DNA packing in filamentous bacteriophages. Annu. Rev. Biophys. Chem., 17, 509-539.
Faruque, S.M. Bin Naser, I., Fujihara, K., Diraphat, P., Chowdhury, N., Kamruzzaman, M., Qadri, F., Yamasaki, S., Ghosh A. N. and Mekalanos, J.J. (2005). Genomic sequence and receptor for the Vibrio cholerae phage KSF-1φ: Evolutionary divergence among filamentousvibriophages mediating lateral gene transfer. J. Bact., 187, 4095-4103.
Gonzalez, M.D., Lichtensteiger, C.A., Caughlan, R. and Vimr, E.R. (2002) Conserved filamentous prophage in Escherichia coli O18:K1:H7 and Yersinia pestis biovar orientalis. J. Bact., 184, 6050-6055.
McLeod, S.M., Kimsey, H.H., Davis, B.M. and Waldor, M.K. (2005). CTXφ and Vibrio cholerae: exploring a newly recognized type of phage–host cell relationship. Molecular Micro., 57, 347–356.
Model, P. and Russel, M. (1988). Filamentous bacteriophage. In: The Bacteriophages, vol. 2, (R. Calendar, Ed.), Plenum Press, New York, pp. 375-456.
Renaudin, J. and Bove, J.M. (1994). SpV1 and SpV4, spiroplasma viruses with circular singlestranded DNA genomes, and their contribution to the biology of spiroplasmas. Adv. Virus Res., 44, 429-463.
Tsuboi, M., Tsunoda, M., Overman, S.A., Benevides, J.M. and Thomas, G.J. Jr. (2010). A structural model for the single-stranded DNA genome of filamentous bacteriophage Pf1. Biochemistry, 49, 1737-1743.
Webb, J.S., Mathew, L. and Kjelleberg, S. (2004). Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bact., 186, 8066-8073.
Contributed by
Day, L.A.
Figures
Figure 1 (Upper) A fanciful diagram to represent all members of the Inoviridae family (from A. Kornberg and T.A. Baker (1991). DNA Replication, 2nd edn. W.H. Freeman, New York; with permission); the gene numbering is for the closely related inoviruses f1, fd and M13 (often referred to, collectively or individually, as Ff phage). (Center) Negative contrast electron micrograph E. coli phage fd showing the differing end morphologies. Bar represents 100 nm; magnification about 180,000 (from C. Gray et al. (1981). J. Mol. Biol., 146, 621627). (Lower) Electron micrograph of innumerable copies of the plectrovirus species Spiroplasma phage SpV1 extruding from the membrane of S. citri; magnification 60,000
(reproduced from Renaudin, J. and Bove, J.M. (1994). Adv. Virus Res., 44, 429463; with permission of Elsevier).
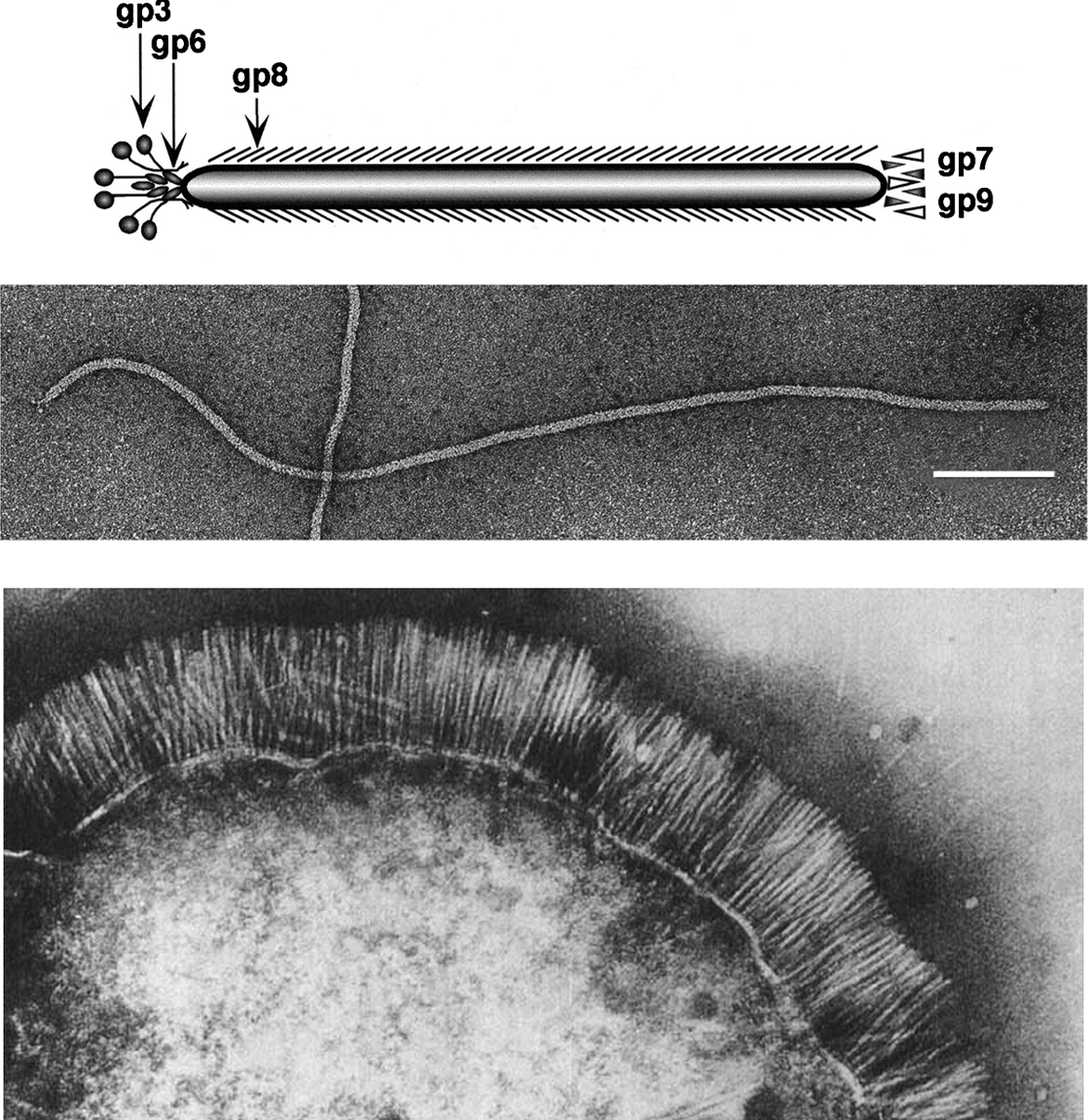
Figure 2 Linear presentations of circular genomes of non-integrative E. coli phage M13 (Upper, 6.4 kb), and integrative Vibrio phage CTX (Lower; 6.9 kb). They are organized in modules of replication genes (red), structural genes (green) and morphogenesis genes (blue). Gene numbers and names are from classical genetic mapping studies Gene 10 (DNA replication) and gene 11 (morphogenesis) are derived translationally from the C-terminal thirds of genes 2 and 1, respectively. Note the loci of the cholera toxin subunits A and B correspond to the locus of g4 in M13 and that the repressor RstR is expressed from the complementary strand.
(Adapted from Faruque, S.M. and Mekalanos, J.J. (2005). J. Bacteriol., 187, 40954103; with permission).