Family:Circoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are not enveloped and exhibit icosahedral symmetry (Figure 1). Ranges reported for virion diameters of chicken anemia virus (CAV), porcine circovirus 1 (PCV1) and beak and feather disease virus (BFDV) are 19.1–26.5 nm, 17–20.7 nm and 12–20.7 nm, respectively. Comparative analysis indicated that the diameters of CAV virions are about 20% greater than those of PCV1 and BFDV, and that CAV exhibits a distinctive surface structure, which is not exhibited by PCV1 and BFDV.
Physicochemical and physical properties
The buoyant densities of virions in CsCl range from 1.33 to 1.37 g cm−3. CAV and PCV1 are resistant to inactivation by treatment at pH 3, and both can withstand incubation at 70°C for 15 min. Both viruses resist treatment with organic solvents such as chloroform, and both show at least partial resistance to sodium dodecyl sulphate.
Nucleic acid
Virions contain a single molecule of circular ssDNA, which ranges from about 1.7 to 2.3 kb in size.
Proteins
The virions of CAV, PCV1 and porcine circovirus 2 (PCV2) are each comprised of one structural protein (capsid protein), for which approximate sizes of 50 kDa (CAV), 30 kDa (PCV1) and 30 kDa (PCV2) have been estimated, respectively. BFDV is reported to contain three proteins, 26.3, 23.7 and 15.9 kDa. The protein composition of virions of the other members of the family Circoviridae is not known, but putative structural proteins have been identified by amino acid similarity searches.
Lipids
Unknown.
Carbohydrates
Unknown.
Genome organization and replication
CAV has a negative sense genome organization, whereas those of the other circoviruses are ambisense. All viruses of the family replicate their genomes using a circular, double strand (ds) replicative form (RF) DNA intermediate, which is produced using host cell DNA polymerases during the S phase of cell division. The RF serves as template for generation of viral ssDNA, probably using the rolling circle replication (RCR) mechanism. Since complex transcription patterns (up to 12 transcripts) have been identified for PCV1 and PCV2, it is also probable that some, if not all, members of the genus Circovirus employ similar transcription strategies. The existence of spliced mRNAs was recently demonstrated for CAV.
Antigenic properties
CAV is antigenically distinct from other circoviruses. PCV1 and BFDV are antigenically different, but PCV1 and PCV2 share common epitopes. The antigenic relationships of avian members of the genus Circovirus are not known.
Biological properties
Circoviruses are host-specific or exhibit a narrow host range, and the majority of those reported infect avian species. Most infections are thought to spread by the fecal–oral route, but vertical transmission has been demonstrated. Circovirus infections are highly prevalent and have widespread geographic distributions. Although subclinical infections are common, circovirus infections are associated with a range of clinical diseases, including infectious chicken anemia, psittacine beak and feather disease, circovirus disease of pigeons, and the postweaning multisystemic wasting syndrome of pigs (PMWS), for which PCV2 has been recognized as an essential component of the disease process. Circovirus infections cause lymphoid depletion and are immunosuppressive.
Genus Circovirus
Type species Porcine circovirus-1
Distinguishing features
Viruses have very similar ambisense genome organizations in which the genes encoding the Rep and CP are divergently arranged on different strands of the ds RF. The intergenic region between the start sites of these genes contains the conserved nonanucleotide motif (TAGTATTAC), located at the apex of a potential stem-loop, at which rolling circle replication (RCR) of the virus DNA initiates.
Virion properties
Morphology
Virions are non-enveloped and show spherical morphology. Electron microscopy studies showed that BFDV and PCV2 have a diameter of about 20.5 nm, and an icosahedral T=1 structure containing 60 CP molecules with dodecahedral symmetry. The virion sizes reported for other circoviruses are similar and appear to depend on the type of negative stain used and on whether measurement is made using negative contrast or thin section electron microscopy.
Physicochemical and physical properties
PCV1 virions have a sedimentation coefficient of 57S and a buoyant density in CsCl of 1.35–1.37 g cm−3.
Nucleic acid
Virions contain covalently closed circular ssDNA. The genomes of PCV1 and PCV2 are the smallest viruses replicating autonomously in mammalian cells (1759 and 1768 nt), and the genomes of the other circoviruses are slightly larger (about 1.7–2 kb). All virus genomes carry a putative stem-loop element with the conserved nonamer (TAGTATTAC) in its apex.
Genome organization and replication
Genomes consist of one ssDNA component approximately 1.7–2 kb in size. Circoviruses possess ambisense genomes with the two major ORFs, from the rep and cap genes, encoded on opposite strands of the replicative dsDNA intermediate (Figure 2). This arrangement creates two intergenic regions, a larger one between the 5′ ends of the two major ORF genes and a shorter one between their 3′ ends. In the case of PCV1 and PCV2, the non-coding region between the initiation codons of the rep and the cap genes comprises the origin of replication. After infection, the ssDNA genome is converted into dsDNA RF, presumably by host enzymes. By analogy to geminiviruses, replication is thought to follow the RCR model and is initiated at the conserved TAGTATT/A sequence. The rep gene of PCV1 and PCV2 expresses two proteins, Rep and Rep′, which are derived from differentially spliced transcripts. Rep is translated from the full-length transcript (PCV1: 312 amino acid residues (aa); PCV2: 314 aa), whereas a spliced transcript encodes truncated and C-terminal frame-shifted Rep′ (PCV1: 168 aa; PCV2: 178 aa). Rep and Rep′ of PCV1 bind to two genomic hexameric repeats, which are located close to the potential stem loop at the origin of replication. Both proteins and binding to the hexamers are essential for initiation of replication. The cap gene of PCV1 and PCV2 encodes the major structural protein Cap (PCV1: 232 aa; PCV2: 233 aa). Cap possesses an arginine-rich N-terminus sequence that is possibly involved in binding to viral DNA, as suggested for PCV1 and PCV2, and BFDV, too. Several smaller ORFs have been identified, but only ORF3 of PCV2 (also identifiable in PCV1) has been recognized as functional, exhibiting an apoptotic activity. The porcine kidney PK-15 cell line can be used successfully for propagation of PCV1 and PCV2. Information regarding other members of the genus Circovirus remains limited.
Antigenic properties
Cap is the main antigenic determinant of PCV1 and PCV2. Both share antigenic epitopes and show cross-reaction. A detailed serological analysis for the avian circoviruses has not yet been performed.
Biological properties
Natural infections with PCV1 and PCV2 appear to be restricted to pigs, while BFDV infections are detected in over 40 species of psittacine birds. Available evidence suggests that viruses are host-specific or have narrow host ranges. The fecal–oral route of transmission is likely, although vertical transmission has been reported in cases of PCV2, BFDV and pigeon circovirus (PiCV).
Species demarcation criteria in the genus
Since the rep genes are highly conserved, species demarcation can be best resolved by phylogenetic comparison of the CP coding sequence (Figure 3). The current suggested criteria demarcating species in the genus are (i) complete genome nt sequence identity <75%, and (ii) CP sequence identity <70%.
List of species in the genus Circovirus
| Beak and feather disease virus |
|
|
| Beak and feather disease virus | [AF080560] | (BFDV) |
| Canary circovirus |
|
|
| Canary circovirus | [AJ301633] | (CaCV) |
| Duck circovirus |
|
|
| Duck circovirus | [AY228555] | (DuCV) |
| Finch circovirus |
|
|
| Finch circovirus | [DQ845075] | (FiCV) |
| Goose circovirus |
|
|
| Goose circovirus | [AJ304456] | (GoCV) |
| Gull circovirus |
|
|
| Gull circovirus | [DQ845074] | (GuCV) |
| Pigeon circovirus |
|
|
| Pigeon circovirus | [AF252610] | (PiCV) |
| Porcine circovirus-1 |
|
|
| Porcine circovirus-1 | [Y09921] | (PCV1) |
| Porcine circovirus-2 |
|
|
| Porcine circovirus-2-pws-PCV | [AF027217] | (PCV2-pws-PCV) |
| Starling circovirus |
|
|
| Starling circovirus | [DQ172906] | (StCV) |
| Swan circovirus |
|
|
| Swan circovirus | [EU056309] | (SwCV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Circovirus but have not been approved as species
| Raven circovirus-4-1131 | [DQ146997] | (RaCV-4-1131) |
Genus Gyrovirus
Type species Chicken anemia virus
Distinguishing features
Chicken anemia virus (CAV), the only member of the genus, can be distinguished from viruses belonging to the genus Circovirus on the basis of its negative sense genome organization. Also, CAV virions are larger than those of other circoviruses, and surface structure is more evident.
Virion properties
Morphology
Virions are non-enveloped and show icosahedral morphology. Electron microscopy studies showed that CAV have a diameter of about 25 nm, with a capsid consisting of 60 subunits (T=1) arranged in 12 pentameric rings.
Physicochemical and physical properties
CAV virions have a sedimentation coefficient of 91S and a buoyant density in CsCl of 1.33–1.35 g cm−3.
Nucleic acid
The genomes of CAV isolates are about 2.3 kb in size and share high sequence identity. Part of the non-transcribed region (NTR) of the genome is G+C-rich and capable of forming stem-loop structures.
Genome organization and replication
Gyroviruses have a negative sense genome organization, with a coding region containing three partially overlapping ORFs (Figure 4). CAV was initially thought to transcribe a single, unspliced mRNA, but recent studies have demonstrated that several spliced transcripts (ca. 2, 1.6, 1.3 and 1.2 kb) are detectable during the life cycle of the virus. NTR contains transcription initiation and termination signals, and a tandemly arranged array of either four or five 19 nt repeats with which promoter–enhancer activity is associated. The circular ssDNA genome is thought to replicate using the RCR mechanism. However, although a semi-conserved nonanucleotide motif, at which RCR initiates in other circular ssDNA replicons, is found within NTR, it is not located at the apex of a potential stem-loop, as is the case with the replicons of some other families of viruses. Three virus proteins are synthesized. VP1 (ca. 52 kDa), encoded by ORF1, is the only structural protein. VP2 (ca. 26 kDa), encoded by ORF2, is a protein phosphatase. VP3 (ca. 14 kDa) encoded by ORF3 is capable of causing apoptosis and has been called “apoptin”. The occurrence within VP1 of two amino acid motifs, which have putative roles in RCR, suggests that this structural protein possesses DNA replication function. Marek’s disease virus-transformed chicken lymphoblastoid MDCC-MSB1 and MDCC-CU147 cell lines can be used successfully for the propagation of CAV isolates.
Antigenic properties
Cell culture isolates of CAV, collected from throughout the world, belong to a single serotype, but isolates can be differentiated by type-specific virus neutralizing monoclonal antibodies.
Biological properties
Although the chicken is the major host, CAV has also been isolated from turkeys, and the detection of virus-specific antibody suggests that Japanese quail may also be susceptible to infection. Vertical (egg) transmission from infected parent chickens results in clinical disease in progeny chicks at 1–2 weeks post-hatching. Clinical signs include anemia and lymphoid depletion. Sub-clinical infections with horizontally acquired virus, probably spread by the fecal–oral route, occur when maternally derived antibody is no longer protective.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Gyrovirus
| Chicken anemia virus |
|
|
| Chicken anemia virus-Cuxhaven-1 | [M55918] | (CAV-Cuxhaven-1) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Gyrovirus but have not been approved as species
None reported.
List of other related viruses which may be members of the family Circoviridae but have not been approved as species
| Cyclovirus-PK5006 | [GQ404844] | (CyCV1-PK5006) |
Note: isolate representative of a group of highly divergent viral genomes identified in humans and chimpanzees.
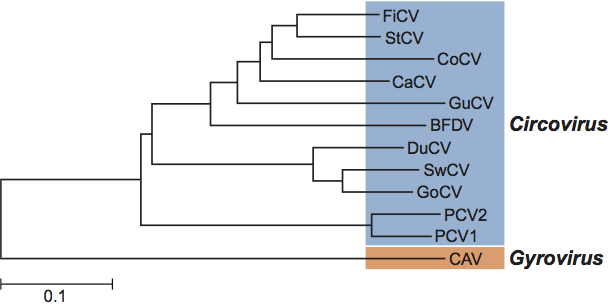
Phylogenetic relationships within the family Circoviridae
Comparison of the Rep proteins of all members of the family Circoviridae reveals that CAV has no close phylogenetic relationship to the species in the genus Circovirus (Figure 5). Similar observations are made when the complete viral genomes are compared.
Similarity with other taxa
Analysis of the genome structure, the origin of replication and the Rep proteins of viruses of the genus Circovirus show similarity to members of the plant virus family Nanoviridae and less pronounced similarity to members of the plant virus family Geminiviridae.
Gyroviruses have features in common with members of the familly Anelloviridae, since they possess circular ssDNA genomes with similar, but not identical, negative sense genome organizations. The non-coding regions of CAV and those of most of the anelloviruses genomes are G+C-rich. The CP of CAV and the putative CPs of anelloviruses possess amino acid sequence motifs that are characteristic of RCR Rep proteins. The proteins encoded by ORF2 in CAV and some anelloviruses contain amino acid sequences that are characteristic of protein tyrosine phosphatases (PTPase), including the motif WX7HX3CXCX5H. Spliced transcripts have been detected for CAV and some anelloviruses, along with peptides able to induce apoptosis in human hepatocellular carcinoma cell lines.
Derivation of names
Circo: from circular conformation.
Gyro: from Latin gyrus, “ring” or “circuit”.
Further reading
Bassami, M.R., Berryman, D, Wilcox, G.E. and Raidal, S.R. (1998). Psittacine beak and feather disease virus nucleotide sequence analysis and its relationship to porcine circovirus, plant circoviruses and chicken anaemia virus. Virology, 249, 453-459.
Crowther, R.A., Berriman, J.A., Curran, W.L., Allan, G.M. and Todd, D. (2003). Comparison of the structures of three circoviruses: chicken anemia virus, porcine circovirus type 2 and beak and feather disease virus. J. Virol., 74, 13036-13041.
Finsterbusch, T. and Mankertz, A. (2009). Porcine circoviruses – small but powerful. Virus Res., 143, 177-183.
Meehan, B.M., Creelan, J.L., McNulty, M.S. and Todd, D. (1997). Sequence of porcine circovirus DNA: affinities with plant circovirus. J. Gen. Virol., 78, 221-227.
Phenix, K.V., Weston, J.H., Ypelaar, I, Lavazza, A., Smyth, J.A., Todd, D., Wilcox, G.E. and Raidal, S.R. (2001). Nucleotide sequence analysis of a novel circovirus of canaries and its relationship to other members of the genus Circovirus of the family Circoviridae. J. Gen. Virol., 82, 2805-2809.
Ritchie, B.W., Niagro, F.D., Lukert, P.D., Steffens, W.L. and Latimer, K.S. (1989). Characterization of a new virus derived from cockatoos with psittacine beak and feather disease. Virology, 171, 83-88.
Schat, K.A. (2009). Chicken anemia virus. Curr. Top. Microbiol. Immunol., 331, 151-183.
Tischer, I., Gelderblom, H., Vetterman, W. and Koch, M.A. (1982). A very small porcine virus with circular single-stranded DNA. Nature, 295, 64-66.
Todd, D., Scott, A.N.J., Fringuelli, E., Shivraprasad, H.L., Gavier-Widen, D. and Smyth, J.A. (2007). Molecular characterization of novel cicroviruses from finch and gull. Avian Pathol., 36, 75-81.
Yuasa, N., Tanigushi, T. and Yoshida, I. (1979). Isolation and some characteristics of an agent inducing anemia in chicks. Avian Dis., 23, 366-385.
Contributed by
Biagini, P., Bendinelli, M., Hino, S., Kakkola, L., Mankertz, A., Niel, C., Okamoto, H., Raidal, S., Teo, C.G. and Todd, D.
Figures
Figure 1 (Left upper) Cryo-electron microscopy image of a particle of an isolate of chicken anemia virus. A structural model comprising 60 subunits (T=1) arranged in 12 trumpet-shaped pentameric rings has been proposed. (Left lower) Cryo-electron microscopy image of a particle of an isolate of porcine circovirus-2. A structural model comprising 60 subunits (T=1) arranged in 12 flat pentameric morphological units has been proposed. (Right) Negative contrast electron microscopy of particles of an isolate of chicken anemia virus (black arrow) and beak and feather disease virus (white arrow), stained with uranyl acetate. Bar=20 nm.
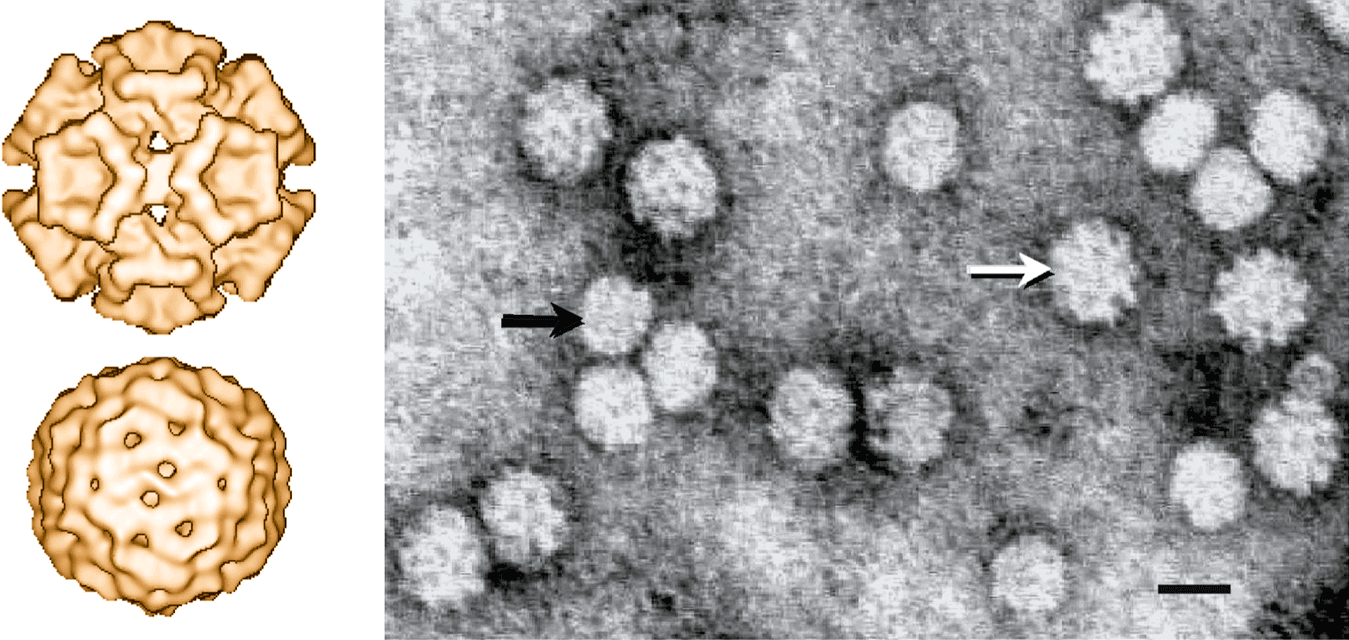
Figure 2 Genome organization of an isolate of porcine circovirus-1.
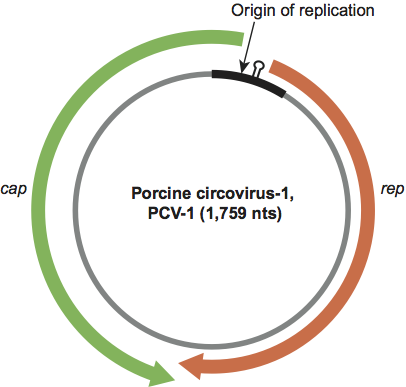
Figure 3 Genus Circovirus: neighbour-joining phylogenetic tree built with CP amino acid sequences.
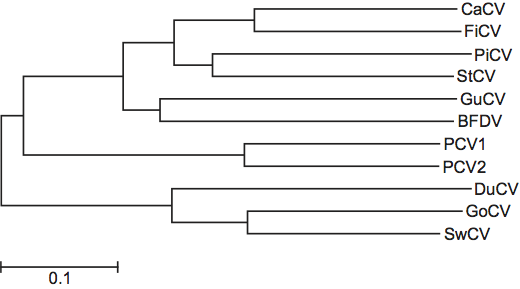
Figure 4 Genome organization of an isolate of chicken anemia virus.
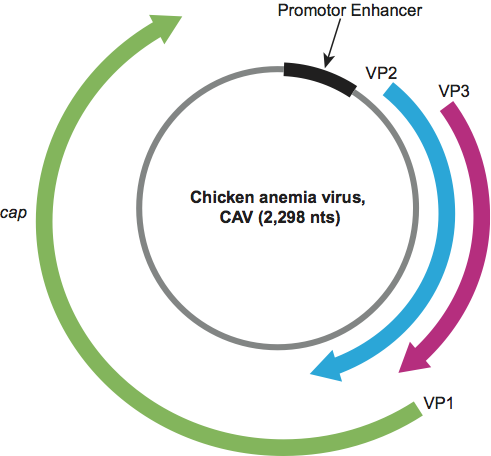
Figure 5 Family Circoviridae: neighbour-joining phylogenetic tree built with Rep amino acid sequences.