Family: Totiviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions in this family are isometric with no lipid or carbohydrate content reported and no surface projections. There is a close similarity in many aspects of the virus particles (Figures 1, 2) of members of the family Totiviridae and the cores of dsRNA viruses of higher organisms.
Physicochemical and physical properties
Virion buoyant density in CsCl is 1.33–1.43 g cm−3. Additional components with different sedimentation coefficients are found in preparations of some viruses in the genus Totivirus. These consist of particles containing satellite or defective dsRNA.
Nucleic acid
Virions contain a single molecule of linear uncapped dsRNA, 4.6–7.0 kbp in size.
Proteins
Virions contain a single major CP, of 70–100 kDa. Virion-associated RNA polymerase activity is present.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genome contains two large, usually overlapping, ORFs: the 5’-proximal ORF encodes the major CP (Gag) and the 3’-proximal ORF encodes an RdRp. Virion-associated RdRp catalyzes in vitroend-to-end transcription of dsRNA to produce mRNA for CP and RdRp, by a conservative mechanism. When provided with viral (+) strand template, the RdRp specifically binds (+) strands and catalyzes (−) strand synthesis to form dsRNA. When provided with viral dsRNA, the RdRp carries out in vitro transcription in a template-specific reaction. The polymerase is usually expressed as a gag-pol-like fusion protein containing two ORFs.
Antigenic properties
Not reported.
Biological properties
These viruses are associated with latent infections of their fungal or protozoan hosts.
Genus Totivirus
Type species Saccharomyces cerevisiae virus L-A
Distinguishing features
Virus replicates in Saccharomyces cerevisiae and supports the replication of one of several satellite dsRNAs (called M dsRNAs) encoding a secreted toxin and immunity to that toxin (killer toxins).
Virion properties
Morphology
Virions are 40 nm in diameter and icosahedral with T=1 with a dimer of the major CP as the asymmetric unit (Figures 1,2). Pores seen in the capsid structure presumably function in the uptake of nucleotides and the release of viral mRNA.
Physicochemical and physical properties
Virion Mr is estimated as 12.3×106. Buoyant density in CsCl is 1.40–1.43 g cm−3 and S20,w is 160–190S. Additional components with different sedimentation coefficients and buoyant densities are present in virus isolates with satellite or defective RNAs. Particles lacking nucleic acid have an S20,w of 98–113S.
Nucleic acid
Virions contain a single linear molecule of uncapped dsRNA (4.6–6.7 kbp). Some virus isolates contain additional satellite dsRNAs which encode “killer” proteins; these satellites are encapsidated separately in capsids encoded by the helper virus genome. Some virus isolates may contain (additionally or alternatively) defective dsRNAs which arise from the satellite dsRNAs; these additional dsRNAs are also encapsidated separately in capsids encoded by the helper virus genome. The complete RNA sequences of Saccharomyces cerevisiae virus L-A (ScV-L-A) (4,579 bp), Saccharomyces cerevisiae virus L-BC (La) (ScV-L-BC) and Ustilago maydis virus H1 (UmV-H1) dsRNAs are available. The positive strand has two large overlapping ORFs; the length of the overlap for ScV-L-A is 130 nt. The first ORF encodes the viral major CP with a predicted size of 76–81 kDa. In the case of ScV-L-A, the two reading frames together encode, via a translational frameshift, the putative RdRp as a fusion protein (analogous to gag-pol fusion proteins of the retroviruses) with a predicted size of 170 kDa. Sites essential for encapsidation, and replication and ribosomal frameshifting have been defined.
Proteins
Virions contain a single major CP of 73–88 kDa. The ScV-L-A Gag removes the 5’ cap structure of host mRNA and covalently attaches it to His154, an activity necessary for expression of viral mRNA. This necessity is relieved by mutation of the host SKI1/XRN1 5’ exoribonuclease specific for uncapped RNAs (e.g., viral (+) strands). RNA polymerase (replicase-transcriptase) is present. In ScV-L-A virions, RNA polymerase occurs as 1–2 molecules of the 170 kDa fusion protein. The Pol domain of the Gag-Pol fusion protein has three single stranded RNA binding activities. The N-terminal 1/4 of Pol is necessary for packaging viral (+) strands and includes one of the RNA binding activities.
Genome organization and replication
ScV-L-A virus has a single 4.6 kbp dsRNA segment with two ORFs (Figure 3). The 5’ ORF is Gag and encodes the major CP that can bind and covalently remove the 5’-cap structure from mRNAs. The 3’ ORF, Pol, encodes the RdRp, and has ssRNA binding activity. Pol is expressed only as a Gag-Pol fusion protein formed by a −1 frameshift in the 130 bp overlap region between the two ORFs. The −1 ribosomal frameshift is produced by a 72 bp region that has a 7 bp slippery site and an essential pseudoknot structure. Covalent linkage of 5’-cap structures (7meGDP) from cellular mRNAs to His154 of Gag is necessary for viral expression, apparently by decoying the Ski1p exoribonuclease from degrading the capless viral (+) strands. The efficiency of frameshifting is critical for viral replication.
Antigenic properties
Virions are efficient immunogens.
Biological properties
Transmission
Virions remain intracellular and are transmitted during cell division, sporogenesis and cell fusion
Host range
ScV-L-A depends for its multiplication on host genes MAK3, MAK10 and MAK31. The MAK3 gene encodes an N-acetyltransferase that acetylates the N-terminus of the major CP and Mak10p and Mak31p are complexed with Mak3p. Reduced levels of 60S ribosomal subunits result in lower levels of ScV-L-A virus and loss of M1 dsRNA, due to poor translation of the viral poly(A)- mRNA. The S. cerevisiae antiviral gene, SKI1, encodes a 5’ to 3’ exoribonuclease specific for uncapped RNAs (such as viral mRNAs). The SKI2, 3, 6, 7, and 8 system blocks expression of non-poly(A) (e.g., viral) mRNAs. Ski2p is an RNA helicase, homologous to nucleolar human homologs. Ski6p is a 3’ to 5’ exoribonuclease involved in 60S ribosomal subunit biogenesis. If an SKI gene is defective, ScV-L-A becomes pathogenic; but only the M dsRNA causes a cytopathogenic effect. Cells become cold sensitive and temperature sensitive for growth.
Species demarcation criteria in the genus
According to the virus species definition, viruses found only in distinct host species are for that reason different species. Totiviruses generally replicate stably within the cell as the cells grow. Different virus strains are expected to segregate relative to each other as the cells grow, whereas different virus species should be stably co-maintained. Viruses of the same species should be similarly affected by host chromosomal mutations. Viruses that can recombine or exchange segments with each other to give viable progeny should be considered the same species. Although these biological criteria are the prime determinants of species, sequence criteria also are used. Less than 50% sequence identity at the protein level generally reflects a species difference. None of the above criteria is absolute, but totiviruses described so far leave little doubt about species demarcation. For example, ScV-L-A and ScV-L-BC are only 30% identical in the 717 aa region of highest similarity. More important, they are stably compatible with each other in the same yeast strain, and respond differently to chromosomal mutations. Mutations resulting in loss of ScV-L-BC do not result in loss of ScV-L-A and vice versa.
List of species in the genus Totivirus
| Saccharomyces cerevisiae virus L-A |
|
|
| Saccharomyces cerevisiae virus L-A | [J04692] | (ScV-L-A) |
| Saccharomyces cerevisiae virus L-BC (La) |
|
|
| Saccharomyces cerevisiae virus L-BC | [U01060] | (ScV-L-BC) |
| Ustilago maydis virus H1 |
|
|
| Ustilago maydis virus H1 | [U01059] | (UmV-H1) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Totivirus but have not been approved as species.
None reported.
Genus Victorivirus
Type species Helminthosporium victoriae virus 190S
Distinguishing features
The RdRp of HvV190S is expressed independent of CP as a separate non-fused virion-associated minor protein. This is also predicted for the RdRps of other members of the genus Victorivirus. The victorivirus CPs are unique among members of the family Totiviridae in having an Ala/Gly/Pro-rich region near their C termini. Victoriviruses replicate in the filamentous fungal host species in which they naturally occur.
Virion properties
Morphology
Virions are isometric, approximately 40 nm in diameter, with icosahedral symmetry (Figure 4). The capsid of HvV190S is made up of 60 asymmetric CP dimers arranged in a T=1 (so-called “T=2” layer). Although the HvV190S capsid shows relatively smoother outer surfaces compared to the ScV-L-A capsid, its quaternary organization is remarkably similar to the ScV-L-A as well as to the cores of the larger dsRNA viruses of plants and animals: the CP-A-subunits cluster around the fivefold axes and CP-B-subunits around the threefold axes.
Physicochemical and physical properties
The buoyant density of HvV190S virions in CsCl is 1.43 g cm−3, and (as its name implies) the sedimentation coefficient is 190S (S20,w, in Svedberg units).
Nucleic acid
The genome of victoriviruses is a single molecule of uncapped dsRNA, consistently sized near 5 kbp, which contains two large ORFs. The upstream ORF codes for the CP and the downstream ORF codes for the RdRp.
Proteins
The HvV190S CP ORF encodes a single primary translation product of 88 kDa (p88). Two additional differently sized forms of CP, designated p83 and p78, are derived from p88 via proteolysis at its C-terminus. In vivo and in vitro phosphorylation studies have indicated that p88 and p83 are phosphoproteins, whereas p78 is not. Two particle types, designated 190S-1 and 190S-2, are present in purified HvV190S preparations. These two particle types are thought to represent different stages in the virus life cycle. The 190S-1 particles contain p88 and p83, and the 190S-2 particles contain p88 and p78. The virion-associated RdRp of HvV190S is present as a separate nonfused minor protein of 92 kDa.
Genome organization and replication
The plus strand of the dsRNA genome of HvV190S encompasses two large ORFs with the 5’-proximal ORF encoding the CP and the 3’-proximal ORF encoding the RdRp. The termination codon of the CP ORF overlaps the initiation codon of the RdRp ORF in the tetranucleotide sequence AUGA (Figure 5). The AUGA overlap region, or a very similar structure, is characteristic of the CP/RdRp junction region of all other members of the genus Victorivirus. ORF 2 in all victoriviruses, with possibly one exception, is in the −1 frame relative to ORF1. The 5’ end of the HvV190S genomic plus strand is uncapped, contains a relatively long (289 nt) 5’ leader with two minicistrons, and is predicted to be highly structured when in single stranded form. These structural features of the 5’ UTR suggest that ORF1 (with its AUG present in suboptimal context according to the Kozak criteria) is translated via a cap-independent mechanism. The HvV190S RdRp is a separate, nonfused virion associated minor protein that is expressed by an internal initiation mechanism; a coupled termination–reinitiation mechanism has been proposed based on current evidence. In in vitro reactions, the RdRp activity associated with HvV190S virions catalyzes end-to-end transcription of dsRNA, by a conservative mechanism, to produce mRNA for CP translation.
Biological properties
Victoriviruses are transmitted intracellularly: vertically during host cell division and sporogenesis and horizontally during cell–cell fusion via hyphal anastomosis between compatible fungal strains. Victoriviruses that infect ascomycetous fungi appear to be largely excluded during ascospore formation.
Species demarcation criteria in the genus
The amino acid sequence identity in pairwise comparisons of either the CP or the RdRp gene product of the nine species is no more than 60%. In addition, each of these species was isolated from a different filamentous fungus, with the exception of one pair. That pair, SsRV1 and SsRV2, is stably co-maintained in the same fungal host and adheres to the 60% maximum identity criterion.
List of species in the genus Victorivirus
| Chalara elegans RNA virus 1 |
|
|
| Chalara elegans RNA virus 1 | [AY561500] | (CeRV1) |
| Coniothyrium minitans RNA virus |
|
|
| Coniothyrium minitans RNA virus | [AF527633] | (CmRV) |
| Epichloe festucae virus 1 |
|
|
| Epichloe festucae virus | [AM261427] | (EfV 1) |
| Gremmeniella abietina RNA virus L1 |
|
|
| Gremmeniella abietina RNA virus L1 | [AF337175] | (GaRV-L1) |
| Helicobasidium mompa totivirus 1-17 |
|
|
| Helicobasidium mompa totivirus 1-17 | [AB085814] | (HmTV1-17) |
| Helminthosporium victoriae virus 190S |
|
|
| Helminthosporium victoriae virus 190S | [U41345] | (HvV190S) |
| Magnaporthe oryzae virus 1 |
|
|
| Magnaporthe oryzae virus 1 | [AB176964] | (MoV1) |
| Sphaeropsis sapinea RNA virus 1 |
|
|
| Sphaeropsis sapinea RNA virus 1 | [AF038665] | (SsRV1) |
| Sphaeropsis sapinea RNA virus 2 |
|
|
| Sphaeropsis sapinea RNA virus 2 | [AF039080] | (SsRV2) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Victorivirus but have not been approved as species
| Botryotinia fuckeliana totivirus 1 | [AM491608] | (BfTV1) |
| Magnaporthe oryzae virus 2 | [AB300379] | (MoV2) |
Genus Giardiavirus
Type species Giardia lamblia virus
Distinguishing features
Giardia lamblia virus (GLV) is a dsRNA virus that replicates in growing Giardia lamblia and is released into the medium without lysing the host cells. It is the only member of the family Totiviridae for which transfection has succeeded. Virions are isometric, 36 nm in diameter (Figure 6).
Virion properties
Morphology
See Figure 6.
Physicochemical and physical properties
Virion buoyant density in CsCl is 1.368 g cm−3.
Nucleic acid
Virions contain a single molecule of dsRNA, 6277 bp in size.
Proteins
Virions contain a single major CP of 98 kDa and a viral RNA polymerase of 190 kDa.
Genome organization and replication
The virus is found in the nuclei of infected G. lamblia. Virus replicates without inhibiting the growth of G. lamblia trophozoites. Virus is also extruded into the culture medium and the extruded virus can infect many virus-free isolates of the protozoan host. There are isolates of the protozoan parasite, however, that are resistant to infection by GLV. A single stranded copy of the viral dsRNA genome is present in infected cells. The concentration of the ssRNA observed during the time course of GLV infection is consistent with a role as a viral replicative intermediate or mRNA. The ssRNA is not capped or polyadenylated.
Biological properties
The virus infects many isolates of G. lamblia, a flagellated protozoan human parasite. The virus does not seem to be associated with the virulence of the parasite. It is not observed in the cyst form of the parasite, and it is not known whether it can be carried through the transformation between cyst and trophozoite. The virus is infectious as purified particles and can infect uninfected G. lamblia.
Species demarcation criteria in the genus
Biological criteria, as for the genus Totivirus, are the prime determinants of species. In addition, less than 50% sequence identity at the protein level generally reflects a species difference. None of these criteria is absolute, but totiviruses described so far leave little doubt about species demarcation.
List of species in the genus Giardiavirus
| Giardia lamblia virus |
|
|
| Giardia lamblia virus | [L13218] | (GLV) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Giardiavirus but have not been approved as species
| Trichomonas vaginalis virus 1 | [U08999] | (TVV1) |
| Trichomonas vaginalis virus 2 | [AF127178] | (TVV2) |
| Trichomonas vaginalis virus 3 | [AF325840] | (TVV3) |
Genus Leishmaniavirus
Type species Leishmania RNA virus 1 - 1
Virion properties
Morphology
Virions are isometric, 33 nm in diameter, with no envelope or surface projections (Figure 7).
Physicochemical and physical properties
Virion buoyant density in CsCl is 1.33 g cm−3.
Nucleic acid
Virions contain a single molecule of linear uncapped dsRNA, 5.3 kbp in size. The complete 5284 nt sequence is available.
Proteins
Virions contain a single major CP of 82 kDa.
Genome organization and replication
The positive strand contains three ORFs (Figure 8). The predicted aa sequence of ORF3 has motifs characteristic of viral RdRp. ORF2 encodes the major CP and overlaps ORF3 by 71 nt, suggesting a +1 translational frameshift to produce a gag-pol-like fusion protein with a predicted size of 176 kDa. Sequencing data support the idea that the abundant ssRNA found in infected cells is the message sense RNA.
Biological properties
Leishmania RNA virus 1- 1 (LRV-1-1) is found in infected Leishmania braziliensis strain CUMC1. Viruses infecting several other strains of L. braziliensis and L. guyanensis are possibly strains of LRV-1-1. A single strain of L. major is known to be infected with LRV-1-1-like virus. The latter is designated LRV-2-1 in order to distinguish it from the viruses infecting new world strains of Leishmania.
Species demarcation criteria in the genus
Biological criteria, as for the genus Totivirus, are the prime determinants of species. In addition, less than 50% sequence identity at the protein level generally reflects a species difference. None of these criteria is absolute, but totiviruses described so far leave little doubt about species demarcation.
List of species in the genus Leishmaniavirus
| Leishmania RNA virus 1 - 1 |
|
|
| Leishmania RNA virus 1 - 1 | [M92355] | (LRV-1-1) |
| Leishmania RNA virus 1 - 2 |
|
|
| Leishmania RNA virus 1 - 2 | [AF230881*] | (LRV-1-2) |
| (Leishmania RNA 2) |
| (LR2) |
| Leishmania RNA virus 1 - 3 |
|
|
| Leishmania RNA virus 1 - 3 |
| (LRV-1-3) |
| Leishmania RNA virus 1 - 4 |
|
|
| Leishmania RNA virus 1 - 4 | [U01899] | (LRV-1-4) |
| (Leishmania B virus) |
| (LBV) |
| Leishmania RNA virus 1 - 5 |
|
|
| Leishmania RNA virus 1 - 5 |
| (LRV-1-5) |
| Leishmania RNA virus 1 - 6 |
|
|
| Leishmania RNA virus 1 - 6 |
| (LRV-1-6) |
| Leishmania RNA virus 1 - 7 |
|
|
| Leishmania RNA virus 1 - 7 | [AF230882*] | (LRV-1-7) |
| Leishmania RNA virus 1 - 8 |
|
|
| Leishmania RNA virus 1 - 8 | [AF230883*] | (LRV-1-8) |
| Leishmania RNA virus 1 - 9 |
|
|
| Leishmania RNA virus 1 - 9 | [AF230884*] | (LRV-1-9) |
| Leishmania RNA virus 1 - 10 |
|
|
| Leishmania RNA virus 1 - 10 | [AF230885*] | (LRV-1-10) |
| Leishmania RNA virus 1 - 11 |
|
|
| Leishmania RNA virus 1 - 11 | [AF230886*] | (LRV-1-11) |
| Leishmania RNA virus 1 - 12 |
|
|
| Leishmania RNA virus 1 - 12 |
| (LRV-1-12) |
| Leishmania RNA virus 2 - 1 |
|
|
| Leishmania RNA virus 2 - 1 | [U32108] | (LRV-2-1) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Leishmaniavirus but have not been approved as species
None reported.
List of other related viruses which may be members of the family Totiviridae but have not been approved as species
| Eimeria brunetti RNA virus 1 | [AF356189] | (EbRV1) |
| Infectious myonecrosis virus | [AY570982] | (IMNV) |
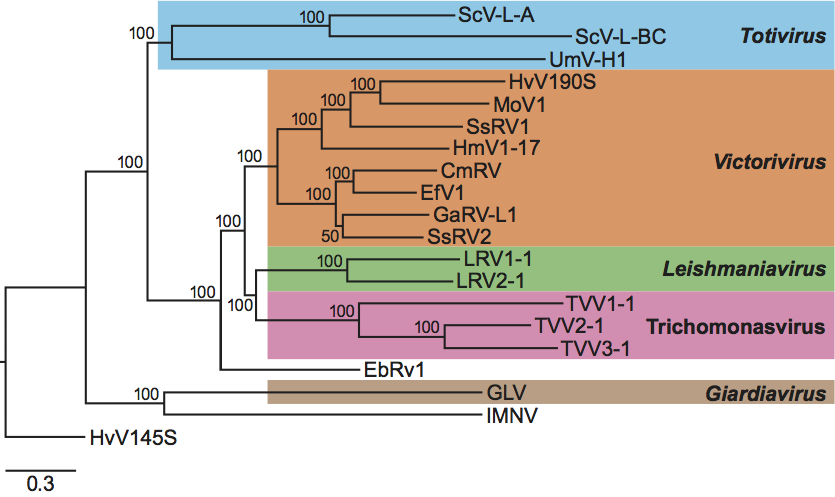
Phylogenetic relationships within the family
Phylogenetic analysis based on amino acid sequences of the RdRps of members of the family Totiviridae (Figure 9) showed that members of each of three recognized genera, Totivirus, Victorivirus and Leishmaniavirus, form separate distinct clusters. The fourth recognized genus (Giardiavirus), with a single species, clusters with the unassigned virus IMNV. It is also clear from such analysis that the Trichomonas vaginalis viruses (TVV1, TVV2 and TVV3) constitute a monophyletic cluster distinguishable from all other viruses in the family, including GLV, the prototype member of the genus Giardiavirus, to which TVVs had been previously assigned on a tentative basis. A proposal to create a new genus for these Trichomonas viruses is under consideration by the ICTV-EC. It is of interest that members of the genus Victorivirus that infect filamentous fungi are more closely related to some protozoan viruses (leishmaniaviruses and Trichomonas viruses as well as the unassigned virus EbRV1) than to members of the genus Totivirus that infect yeast and smut fungi. This further justifies the placement of viruses that infect filamentous fungi, which were once included in the genus Totivirus, in their own separate genus, Victorivirus.
Similarity with other taxa
The RdRp of each virus has the consensus sequences typical of the RdRps of (+) ssRNA and dsRNA viruses. The capsid structures have the “T=2” structure with 120 monomers, typical of the cores of all dsRNA viruses but of no other viruses. The replication and transcription strategies of the L–A virus resemble those of other dsRNA viruses.
Derivation of names
Giardia: derived from the name of the host.
Leishmania: derived from the name of the host.
Toti: from Latin totus, “whole” or “undivided”.
Victori: derived from the species name of the host, Helminthosporium victoriae.
Further reading
Dinman et al., 1991 J.D. Dinman, T. Icho, R.B. Wickner, A-1 ribosomal frameshift in a double stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl Acad. Sci., U S A. 88 (1991) 174–178.
Fujimura et al., 1992 T. Fujimura, J.C. Ribas, A.M. Makhov, R.B. Wickner, Pol of Gag-Pol fusion protein required for encapsidation of viral RNA of yeast L-A virus. Nature. 359 (1992) 746–749.
Ghabrial, 2008 S.A. Ghabrial, Totiviruses. In: . Elsevier, Oxford2008163–174.
Ghabrial and Nibert, 2009 S.A. Ghabrial, M.L. Nibert, Victorivirus, a new genus of fungal viruses in the family Totiviridae. Arch. Virol. 154 (2009) 373–379.
Icho and Wickner, 1989 T. Icho, R.B. Wickner, The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J. Biol. Chem. 264 (1989) 6716–6723.
Kang et al., 2001 J. Kang, J. Wu, J.A. Bruenn, C. Park, The H1 double-stranded RNA genome of Ustilago maydis virus-H1 encodes a polyprotein that contains structural motifs for capsid polypeptide, papain-like protease, and RNA-dependent RNA polymerase. Virus Res. 76 (2001) 183–189.
Koltin, 1988 Y. Koltin, Y. Koltin, M. Leibowitz, The killer systems of Ustilago maydisViruses of Fungi and Simple Eukaryotes. In: Y. Koltin, M. Leibowitz, Viruses of Fungi and Simple Eukaryotes. Marcel Dekker, New York1988209–243.
Naitow et al., 2002 H. Naitow, J. Tang, M. Canady, R.B. Wickner, J.E. Johnson, L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism. Nat. Struct. Biol. 9 (2002) 725–728.
Stuart et al., 1992 K.D. Stuart, R. Weeks, L. Guilbride, P.J. Myler, Molecular organization of Leishmania RNA virus. Proc. Natl Acad. Sci., U S A. 89 (1992) 8596–8600.
Tercero and Wickner, 1992 J.C. Tercero, R.B. Wickner, MAK3 encodes an N-acetyltransferase whose modification of the L-A gag N-terminus is necessary for virus particle assembly. J. Biol. Chem. 267 (1992) 20277–20281.
Wang and Wang, 1991 A.L. Wang, C.C. Wang, Viruses of the protozoa. Ann. Rev. Microbiol. 45 (1991) 251–263.
White and Wang, 1990 T.C. White, C.C. Wang, RNA dependent RNA polymerase activity associated with the double-stranded RNA virus of Giardia lamblia. Nucleic Acids Res. 18 (1990) 553–559.
Wickner, 1996 R.B. Wickner, Double-stranded RNA viruses of yeast. Microbiol. Rev. 60 (1996) 250–265.
Wickner, 2007 R.B. Wickner, D.M. Knipe, P.M. Howley, Viruses and prions of yeasts, fungi and parasitic microorganismsFields Virology. In: D.M. Knipe, P.M. Howley, Fields Virology. Lippincott, Williams and Wilkins, Philadelphia, PA2007737–768.
Contributed by
Wickner, R.B., Ghabrial, S.A., Nibert, M.L., Patterson, J.L. and Wang, C.C.
Figures
Figure 1 (Left) Reconstruction of the atomic resolution structure of the Saccharomyces cerevisiae virus L-A (ScV-L-A) virion. (Center) Schematic representation of a T=1 capsid structure. (Right) The ScV-L-A particle viewed down an icosahedral two-fold axis with the C positions traced. Red Gag molecules contact the three-fold axes, but not the two-fold or five-fold axes. Purple molecules surround the five-fold axes and contact the two-fold axes. Thus, two kinds of Gag molecules with identical covalent structure are in distinct environments in the viral particle.
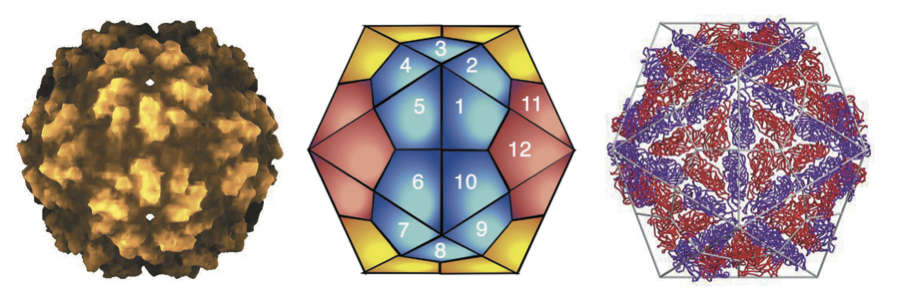
Figure 2 Cryo-electron microscopic reconstruction of Saccharomyces cerevisiae virus L-A (ScV-L-A) at 16 resolution. The view shown is along a five-fold axis of the icosahedral particles. The virions show the T=2 symmetry found in the cores of all dsRNA viruses.
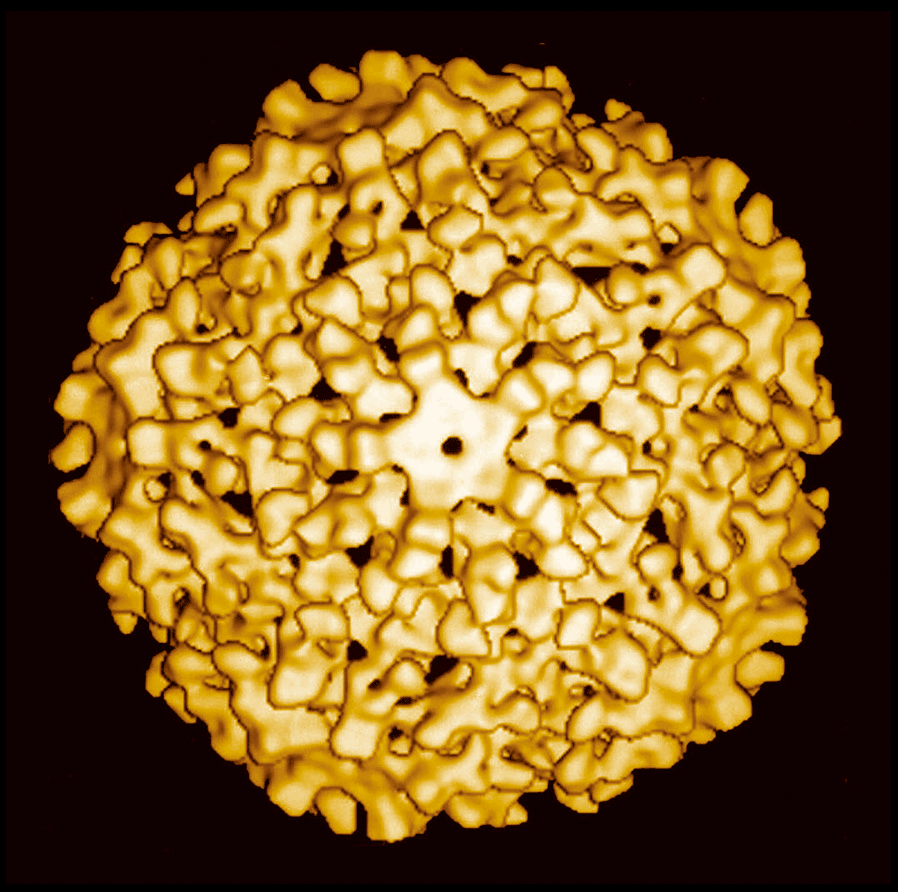
Figure 3 Genome organization of Saccharomyces cerevisiae virus L-A (ScV-L-A). The virion-associated RNA polymerase catalyzes in vitro end-to-end transcription of dsRNA by a conservative mechanism to produce mRNA for CP. In the case of ScV-L-A, all of the positive strand transcripts are extruded from the particles. The positive strand of satellite RNA M1, or deletion mutants of L-A or M1, on the other hand, often remain within the particle where they are replicated to produce two or more dsRNA molecules per particle (headful replication). The positive ssRNA of ScV-L-A is the molecule encapsidated to form progeny virus particles. The encapsidation signal on ScV-L-A or M1 positive sense ssRNA is a 24 nt stem-loop sequence located 400 nt from the 3 end in each case. The Gag protein must be acetylated (by the cellular Mak3p) for assembly and packaging to proceed. These particles have a replicase activity that synthesizes the negative strand on the positive strand template to produce dsRNA, thus completing the replication cycle. Replication requires an internal site overlapping the packaging signal, and a specific 3-end sequence and secondary/tertiary structure. Virions accumulate in the cytoplasm.
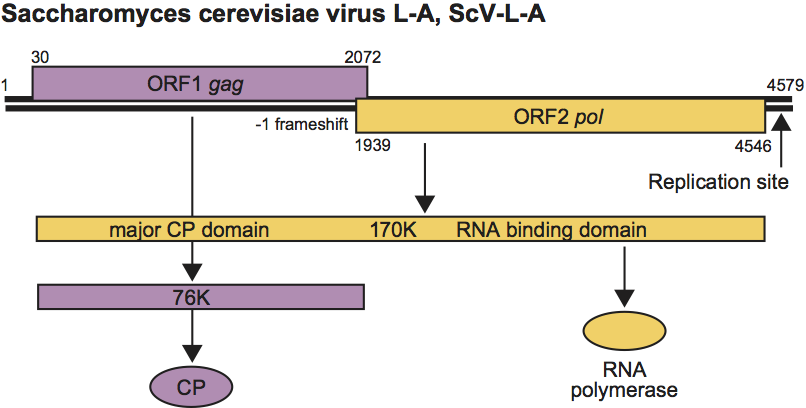
Figure 4 Negative contrast electron micrograph of an isolate of Helminthosporium victoriae virus 190S, the type member of the genus Victorivirus.
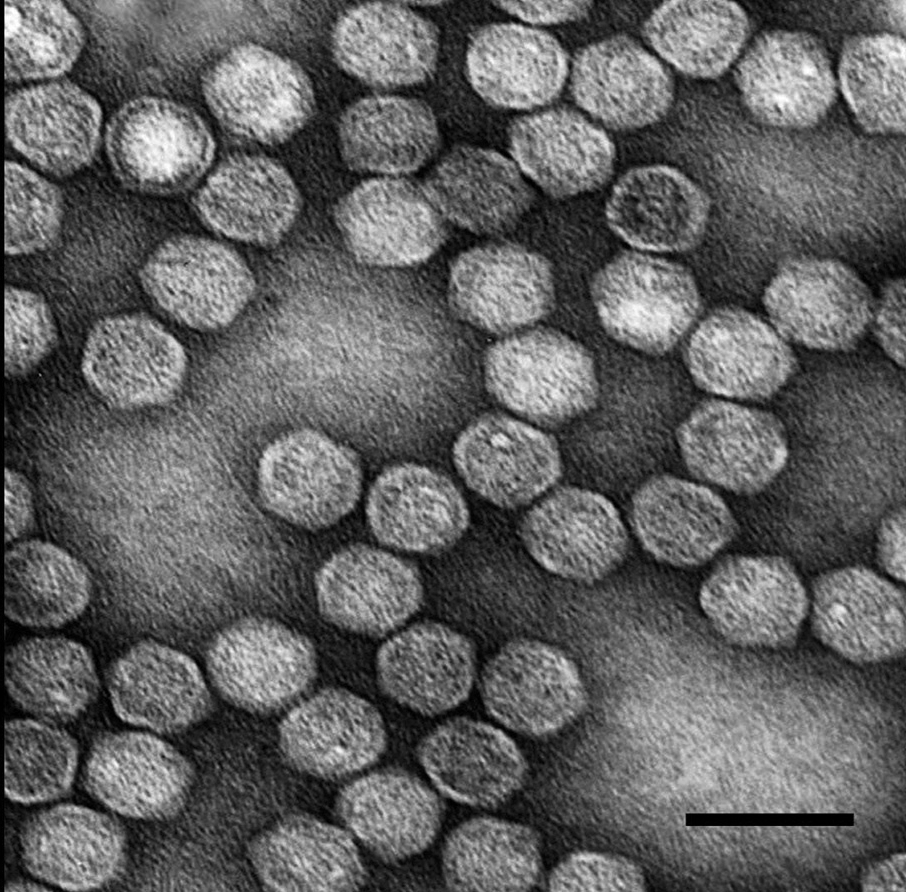
Figure 5 Genome organization of Helminthosporium victoriae virus 190S. The genomic plus strand includes two large, overlapping ORFs, with the 5 one (ORF1) encoding the CP and the 3 one (ORF2) encoding the RdRp. The stop codon of ORF1 overlaps the start codon of ORF2 in the tetranucleotide sequence AUGA.
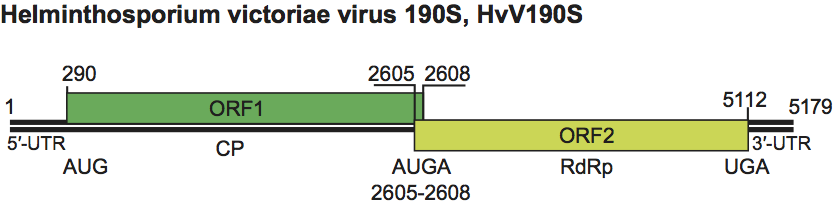
Figure 6 Negative contrast electron micrograph of particles of an isolate of Giardia lamblia virus. TMV (rod-shaped) is included as an internal size marker. The bar represents 100 nm.
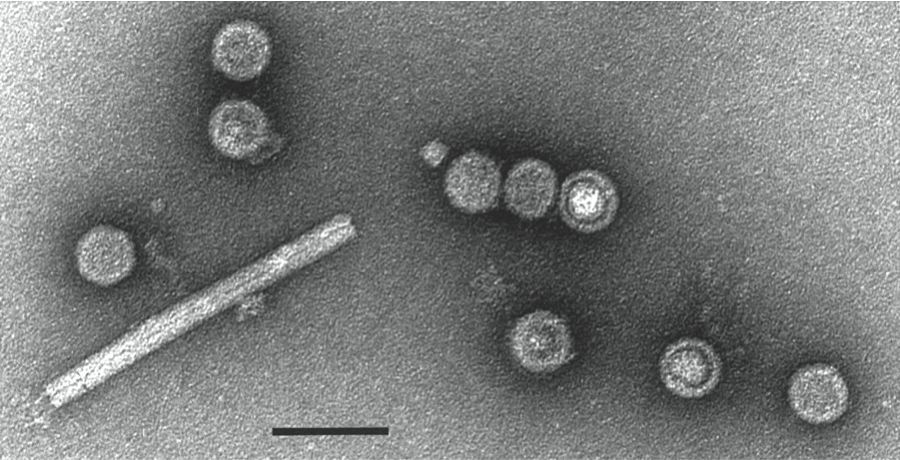
Figure 7 Negative contrast electron micrograph of particles of an isolate of Leishmania RNA virus 1 - 1. The bar represents 100 nm.
Figure 8 Genome organization of Leishmania RNA virus 1 - 1 (LRV-1-1).
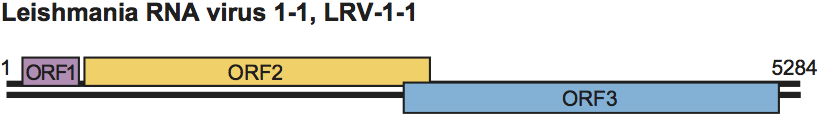
Figure 9 Phylogenetic relationships among members of the family Totiviridae. A Bayesian tree was derived from the RdRp ORF amino acid sequences of each analyzed virus. The multiple sequence alignment was generated using MUSCLE v3.7 without Gblocks curation; the phylogenetic tree was generated using MrBayes v3.1.2 (10,000 generations with sampling every 100 and first 100 generations discarded); and the tree was rendered using TreeDyn v198.3. All four steps were performed using the la carte option at www.phylogeny.fr/ with other default settings. The tree was refined for presentation using FigTree v1.3.1 obtained from http://tree.bio.ed.ac.uk/software/figtree/. The tree is rooted on outgroup virus HvV145S (Helminthosporium victoriae virus 145S; GenBank accession no. YP_052858) from the family Chrysoviridae, and the scale bar indicates the number of substitutions per position in the alignment. Approved and probable members of the family Totiviridae included in the tree are (GenBank accession no. in parenthesis): ScV-L-A, Saccharomyces cerevisiae virus L-A (NP_620495.1); ScV-L-BC, Saccharomyces cerevisiae virus L-BC (NP_042581.1); UmV-H1, Ustilago maydis virus H1 (NC_003823.1); HvV190S, Helminthosporium victoriae virus 190S (AAB94791.2); MoV1, Magnaporthe oryzae virus 1 (YP_122352.1); HmV1-17, SsRV1, Sphaeropsis sapinea RNA virus 1 (NP_047558.1); Helicobasidium mompa totivirus 1-17 (NP_898833.1); CmRV, Coniothyrium minitans RNA virus (YP_392467.1); EfV1, Epichloe festucae virus 1 (CAK02788.1); GaRV-L1, Gremmeniella abietina RNA virus L1 (AAK11656.1); SsRV2, Sphaeropsis sapinea RNA virus 2 (NP_047560.1); LRV1-1, Leishmania RNA virus 1 - 1 (NP_041191.1); LRV2-1, Leishmania RNA virus 2 - 1 (NP_043465.1); TVV1-1, Trichomonas vaginalis virus 1-1 (AAA62868.1); TVV2-1, Trichomonas vaginalis virus 2-1 (AAF29445.1); TVV3-1, Trichomonas vaginalis virus 3-1 (NP_659390.1); EbRV1, Eimeria brunetti RNA virus 1 (NP_108651.1); GLV, Giardia lamblia virus (NP_620070.1); and IMNV, penaeid shrimp infectious myonecrosis virus (YP_529549.1). Viruses are clustered and labeled as follows: genus Totivirus (cyan), genus Victorivirus (orange), genus Leishmaniavirus (green), proposed new genus of Trichomonas viruses (magenta), and genus Giardiavirus (brown). Probable family members EbRV1 and IMNV are indicated in gray.