Family: Picobirnaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Picobirnavirus
Type species Human picobirnavirus
Virion properties
Morphology
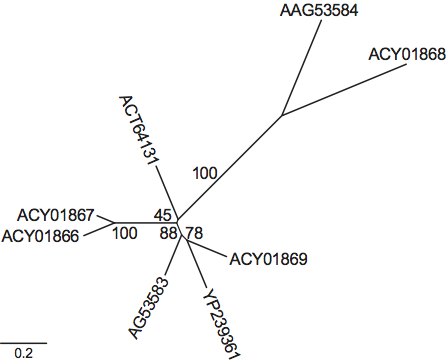
Virions are isometric, non-enveloped, 33–37 nm in diameter. Virus particles have a spherical triacontahedral (30-sided) organization (Figure 1), with a unique capsid layer surrounding the genomic dsRNA segments. Owing to the absence of a cell culture system for propagating picobirnaviruses, knowledge of virion protein composition comes from virion-like particles produced by recombinant expression of the capsid precursor gene (ORF 3) of segment 1 of rabbit picobirnavirus (PBV). The particle is made of 60 symmetric CP dimers. The structure reveals an intricate interface with the N-terminal residues exchanged between the two subunits of the dimer. Two domains can be recognized in CP, namely a shell domain and a projection domain that forms the protrusions that stand out in the three-dimensional reconstruction (Figure 1, left).
Physicochemical and physical properties
Virion buoyant density in CsCl is in the range of 1.38–1.4 g cm−3.
Nucleic acid
Virions contain two unrelated, linear dsRNA segments (named 1 and 2). The larger segment 1 is about 2.4–2.6 kbp long and possesses three ORFs. The smaller segment 2 is 1.5–1.9 kbp long and is monocistronic. It encodes the viral RNA-dependent RNA polymerase (RdRp).
Lipids
Not present.
Genome organization and replication
Only two complete sequences of genome segment 1, from human and rabbit PBVs, are available in sequence databases. The genome organization of segment 1 of human PBV is illustrated in Figure 2. Segment 1 possesses two large ORFs of 224 (ORF2) and 590 (ORF3) codons preceded by a short one (ORF1) of 39 codons. The three ORFs overlap at eight (ORF1–ORF2 junction) and one (ORF2–ORF3 junction) nucleotides (nt), in each case excluding the stop codon of the preceding ORF. While the functionality of ORF1 is unclear and ORF2 encodes a protein with an unknown function, more information is available for ORF3. This encodes the precursor of CP. For rabbit PBV, ORF3 expression results in the synthesis of a precursor that is autocatalytically cleaved to generate a large peptide and mature CP (Figure 3). Cleavage occurs between amino acid residues 65 and 66. Segment 2 encodes a RdRp, whose core component contains the canonical A–B–C motif arrangement of the palm subdomain present in conventional nucleic acid polymerases.
Antigenic properties
No data are available.
Biological properties
Natural host range
PBVs are widely distributed geographically among humans and mammals in general, and have also been reported in birds and reptiles. PBVs have been identified from fecal specimens owing to large amounts of virus occasionally shed through feces. The viruses have also been detected in raw sewage samples.
Pathology
The pathogenicity of PBVs has not been established. Studies conducted with immunocompromised persons suggest that PBVs are opportunistic pathogens that may cause diarrhea. PBVs have been detected in stool samples from children with diarrhoea as well as in immunocompromised patients, and they have also been detected in individuals lacking symptoms of gastroenteritis.
Transmission
Genome sequencing of PBVs originating from different species suggests effective animal-to-animal transmission. The data do not provide evidence of virus clusters specific to a host species.
Species demarcation criteria in the genus
Only two sequences of capsid protein are yet available in databases. The capsid protein of rabbit picobirnavirus displays 25% amino acid sequence identity with its human counterpart, showing that they are phylogenetically distant. The PBVs have been divided into two genogroups defined on the basis of nucleotide sequence similarities in the genomic segment that encodes the RdRp (segment 2). The reference viruses for genogroups I and II are HY005102 and 1-CHN-97, respectively. Genogroup II PBVs are identified on a less frequent basis than genogroup I viruses. Segment 2 of new human (VS10) and bovine (bovine/RUBV-P/IND/2005) isolates showed 55–65% identity at the amino acid sequence level to human prototype strains HY005102 and 1-CHN-97. These observations suggest that PBVs exhibit a high level of genetic diversity that is not reflected by the two prototype viruses. Viruses are distinguished on the basis of their host, the country in which they were identified and the year of isolation.
List of species in the genus Picobirnavirirus
| Human picobirnavirus |
|
|
| Human picobirnavirus - Hy005102 | [RNA 1: AB186897, RNA 2: AB186898] | (human PBV-Hy005102) |
| Rabbit picobirnavirus |
|
|
| Rabbit picobirnavirus - R5-9 | [RNA 1: AJ244022] | (rabbit PBV-R5-9) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Picobirnavirirus but have not been approved as species
| Bovine picobirnavirus | [RNA 2: GQ221268] |
|
| Chicken picobirnavirus |
|
|
| Dog picobirnavirus | [RNA 2: FJ164032] |
|
| Foal picobirnavirus |
|
|
| Giant anteater picobirnavirus |
|
|
| Guinea pig picobirnavirus |
|
|
| Hamster picobirnavirus |
|
|
| Porcine picobirnavirus | [RNA 2: EU104358] |
|
| Rat picobirnavirus | [RNA 2: EU814972] |
|
| Snake picobirnavirus | [RNA 2: EU814971] |
|
Phylogenetic relationships within the family
Phylogenetic relationships within the family are shown in Figure 4.
Similarity with other taxa
The crystal structure of a partitivirus, Penicillium stoloniferum virus F (PsV-F, also a dsRNA virus) also features 60 symmetric dimers, with an overall organization similar to that of the PBV particle. The PsV-F capsid appears slightly smaller than its PBV counterparts. Also, instead of the projections observed in PBV, it exhibits an “arch” that protrudes at the center of each dimer. In contrast to what is observed in PBVs, no maturation cleavage occurs during partitivirus particle formation.
There is no similarity between PBVs and birnaviruses, as assessed from differences in their genome organizations, capsid structures and encoded proteins.
Derivation of name
Picobirna: from Greek pico, “small”; Latin prefix bi, “two”, signifies the bisegmented nature of the viral genome as well as the presence of dsRNA; and rna, abbreviation of ribonucleic acid, indicating the nature of the genome.
Further reading
Journals and books
Bányai, K., Martella, V., Bogdán, A., Forgách, P., Jakab, F., Meleg, E., Bíró, H., Melegh, B. and Szucs, G. (2008). Genogroup I PBVs in pigs: evidence for genetic diversity and relatedness to human strains. J. Gen. Virol., 89, 534-539.
Carruyo, G.M., Mateu, G., Martínez, L.C., Pujol, F.H., Nates, S.V., Liprandi, F. and Ludert, J.E. (2008). Molecular characterization of porcine picobirnaviruses and development of a specific reverse transcription-PCR assay. J. Clin. Microbiol., 46, 2402-2405.
Duquerroy, S., Da Costa, B., Henry, C., Vigouroux, A., Libersou, S., Lepault, J., Navaza, J., Delmas, B. and Rey, F.A. (2009). The picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. EMBO J., 28, 1655-1665.
Ghosh, S., Kobayashi, N., Nagashima, S. and Naik, T.N. (2009). Molecular characterization of full-length genomic segment 2 of a bovine picobirnavirus (PBV) strain: evidence for high genetic diversity with genogroup I PBVs. J. Gen.Virol., 90, 2519-2524.
Green, J., Gallimore, C.I., Clewley, J.P. and Brown, D.W. (1999). Genomic characterisation of the large segment of a rabbit picobirnavirus and comparison with the atypical picobirnavirus of Cryptosporidium parvum. Arch. Virol., 144, 2457-2465.
Pereira, H.G., Flewett, T.H., Candeias, J.A. and Barth, O.M. (1988). A virus with a bisegmented double-stranded RNA genome in rat (Oryzomys nigripes) intestines. J. Gen. Virol., 69, 2749-2754.
Rosen, B.I., Fang, Z.Y., Glass, R.I. and Monroe, S.S. (2000). Cloning of human picobirnavirus genomic segments and development of an RT-PCR detection assay. Virology, 277, 316-329.
Van Leeuwen, M., Williams, M.M., Koraka, P., Simon, J.H., Smits, S.L. and Osterhaus, A.D. (2010). Human picobirnaviruses identified by molecular screening of diarrhea samples. J. Clin. Microbiol., 48, 1787-1794.
Victoria, J.G., Kapoor, A., Li, L., Blinkova, O., Slikas, B., Wang, C., Naeem, A., Zaidi, S. and Delwart, E. (2009). Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J. Virol., 83, 4642-4651.
Wakuda, M., Pongsuwanna, Y. and Taniguchi, K. (2005). Complete nucleotide sequences of two RNA segments of human picobirnavirus. J. Virol. Methods, 126, 165-169.
Websites
DPVWEB: http://www.dpvweb.net/notes/showfamily.php?family=Picobirnaviridae
WIKIMEDIA: http://species.wikimedia.org/wiki/Picobirnaviridae
Contributed by
Delmas, B.
Figures
Figure 1 Structure of the PBV particle. (Left) Surface rendering from a three-dimensional reconstruction viewed down a slightly misoriented five-fold axis. Note the presence of 60 dimeric protrusions. (Middle) Diagram representing a triacontahedron, a convex polyhedron made of 30 rhombic faces or diamond tiles. (Right) Triacontahedral design of the particle, with each of the tiles formed by two CP dimers colored differently
(Courtesy of J. Lepault and S. Duquerroy for the left and right panels, respectively.)
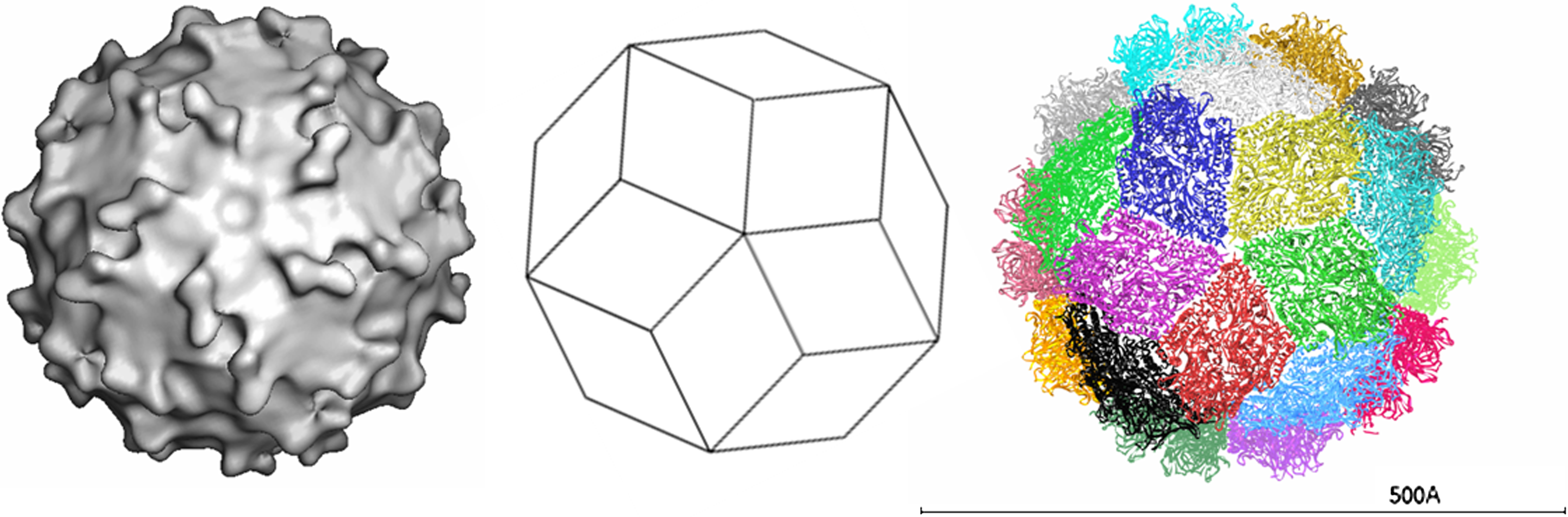
Figure 2 Schematic representation of the gene arrangement in genome segments 1 and 2 of human picobirnavirus. Segment 1 possesses three ORFs. ORF3 encodes the CP precursor. Segment 2 encodes an RNA-dependent RNA polymerase (RdRp). Numbers indicate the positions and lengths of the ORFs.
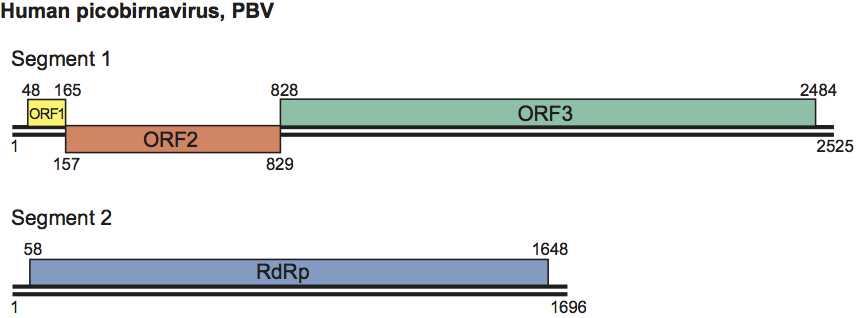
Figure 3 Schematic representation of the gene arrangement in genome segment 1 of rabbit PBV and processing of the CP precursor. Numbers indicate the nucleotide the amino acid positions (the latter in bold type).
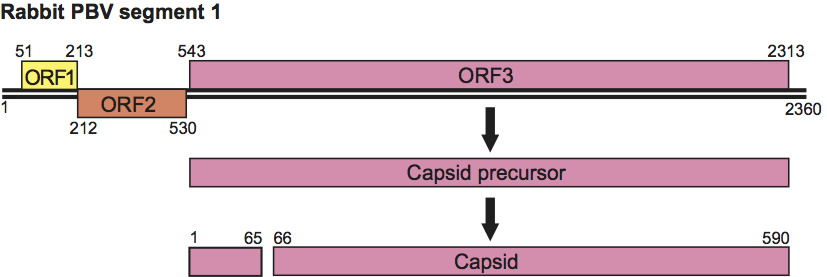
Figure 4 A distance tree representing the phylogenetic relationships among the RNA-dependent RNA polymerases (RdRps) from genetic clusters in the family. The tree was constructed using the software Mega4 and the NJ algorithm, with the option complete deletion, which ensures that only the positions represented in all sequences of the ClustalW multiple alignment were used for the tree calculation. Numbers indicate bootstrap percentages obtained for each node. Sequences are represented by their protein accession numbers. All derive from human picobirnavirus isolates except ACT64131, which corresponds to a bovine isolate. Two genogroups have been defined thus far on the basis of RdRp sequences. The prototype strains are HY005102 (YP_239361) and 1-CHN-97 (AAG53583) for genotype 1 and 4-GA-91 (AAG53584) for genotype 2.