Family: Partitiviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are isometric, non-enveloped, 30–43 nm in diameter (Figure 1). Each virion contains 120 copies of a single capsid protein (CP) arranged as 60 dimers with T=1 icosahedral symmetry. Dimeric surface protrusions are frequently observed on viral capsids.
Nucleic acid
Virions contain two unrelated, linear dsRNA segments (1.4–2.4 kbp in size). The two segments of the individual viruses are usually of similar size. The dsRNA segments are packaged in separate particles.
Proteins
There is a single major CP and a separately translated RdRp. Virion-associated RNA polymerase activity is present.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genome consists of two linear dsRNA segments; the smaller usually codes for the CP and the larger usually codes for the virion-associated RNA polymerase. Each dsRNA is monocistronic. In vitro transcription/replication occurs by a semi-conservative mechanism. Virions accumulate in the cytoplasm.
Antigenic properties
Virions are efficient immunogens. No serological relationships between the fungal viruses and the plant viruses in the family Partitiviridae have been detected.
Biological properties
The viruses are associated with latent infections of their fungal, protozoan and plant hosts. There are no known natural vectors. The fungal partitiviruses are transmitted intracellularly during cell division, hyphal anastomosis and sporogenesis. In some ascomycetes (e.g. Gaeumannomyces graminis), virus is usually eliminated during ascospore formation. Experimental transmission of purified fungal partitiviruses has been reported by transfection of virions into fungal protoplasts. The plant cryptoviruses are transmitted by ovule and by pollen to the seed embryo. There is no graft transmission and apparently no cell-to-cell transport, except at cell division; seed transmission is the only known mode for the transmission of alpha- and betacryptoviruses.
Genus Partitivirus
Type species Atkinsonella hypoxylon virus
Distinguishing features
Members of the genus Partitivirus infect only fungi and are transmitted intracellularly during cell fusion, cell division and sporogenesis. All members have two dsRNA segments of similar sizes that are individually encapsidated in separate particles.
Virion properties
Morphology
The structures of Penicillium stoloniferum viruses F and S (PsV-F and PsV-S) have been determined by X-ray crystallography and/or electron cryomicroscopy, respectively (Figure 2). The outer diameter of the capsid is about 35–40 nm. The most prominent features of the capsid are 60 arch-like protrusions that decorate a spherical shell (Figure 2A–C). Each particle contains 120 CP molecules arranged with icosahedral symmetry. The tertiary structure of each CP molecule consists of two distinct domains, one of which forms the continuous, 3-nm thick, capsid, and the other that, with the corresponding domain of a neighboring CP, comprises the arch (Figure 1E–F). The asymmetric unit of the icosahedron consists of two related CP molecules, CPA and CPB. The 60 CPA molecules are organized as flower-like pentamers, each centered about one of the 12 vertices of the icosahedral capsid (Figure 2A). The 60 CPB molecules are arranged as trimers at the twenty icosahedral 3f axes (Figure 2A). The CPA-CPB dimer, defined by two monomers that form an arch, is most stable based on buried surface areas and likely functions as the assembly precursor (Figure 2E–F). The two CP molecules in this dimer assume nearly identical conformations and are related by almost-perfect local 2f symmetry. Each dimer is stabilized by extensive structure swapping between the monomers and the additional intersubunit interactions mediated by the arch tip. Unlike the assembly pathway that has been described for reoviruses and other larger dsRNA viruses, in PsV-F and PsV-S assembly is likely to proceed from dimers of CP dimers, each of which adopts a striking, smooth-edged diamond shape (Figure 2D). The N-terminal region (ca. 40 aa) of the CP polypeptide extends radially inwards and interacts with the dsRNA viral genome. Given the large number of basic residues within this region, it may participate in RNA packaging during particle assembly, and/or play a role during viral transcription. Small pores in the capsid shell at the icosahedral 5f and 3f axes may facilitate the export of RNA transcripts. The dsRNA genome appears as concentric layers in icosahedrally averaged maps.
Physicochemical and physical properties
Virion Mr is estimated to range from 6 to 9 ×106. S20,w values range from 101 to 145S. Particles lacking nucleic acid have an S20,w of 66–100S. Virion buoyant density in CsCl is 1.29–1.30 and 1.34–1.36 g cm−3 for particles without and with nucleic acid, respectively. Components with other density values and sedimentation rates are found in preparations of some viruses and are believed to be replicative intermediates. These consist of particles containing ssRNA and particles with both ssRNA and dsRNA. Virus purification is usually carried out at neutral pH.
Nucleic
Virions contain two unrelated, linear dsRNA segments, 1.4–2.4 kbp in size, which are separately encapsidated. The dsRNA segments of the individual viruses are of similar size. Additional dsRNA segments (satellite or defective) may be present.
Proteins
There is a single major CP sized 44–77 kDa. The sizes of the RNA-dependent RNA polymerase, as deduced from nucleotide sequence analysis, range from 60 to 87 kDa. Virion-associated RNA polymerase activity is present.
Genome organization and replication
Atkinsonella hypxylon virus (AhV), an isolate of the type species of the genus Partitivirus, has a bipartite genome consisting of dsRNA1 and dsRNA2 (Figure 3). Each is monocistronic: dsRNA1 codes for the RdRp and dsRNA2 codes for the major CP. The virion-associated RdRp catalyzes in vitro end-to-end transcription of each dsRNA to produce mRNA by a semi-conservative mechanism. Virions accumulate in the cytoplasm. A model for the replication strategy of PsV-S is shown in Figure 4.
Species demarcation criteria in the genus
The criteria to differentiate species in the genus are:
- Host species in which the viruses naturally occur; partitiviruses lack natural vectors and they are confined to the fungal host species from which they were first isolated
- Sizes of dsRNA segments and encoded proteins
- Protein sequence similarity. Amino acid sequence similarity >40% between RdRps of viruses from different species in the same phylogenetic cluster and <40% between members of species in different clusters (see Figure 8; phylogenetic tree)
List of species in the genus Partitivirus
| Agaricus bisporus virus 4 |
|
|
| Agaricus bisporus virus 4 |
| (AbV-4) |
| Aspergillus ochraceous virus 1 |
|
|
| Aspergillus ochraceous virus 1-FA0611 | [EU118277, EU118278] | (AoV1-FA0611) |
| Atkinsonella hypxylon virus |
|
|
| Atkinsonella hypxylon virus – 2H | [L39125, L39126, L39127] | (AhV-2H) |
| Ceratocystis resinifera virus 1 |
|
|
| Ceratocystis resinifera virus 1 | [AY603051, AY603052] | (CrV1) |
| Discula destructiva virus 1 |
|
|
| Discula destructiva virus 1 – 247 | [AF316992, AF316993, AF316994, AF316995] | (DdV1-247) |
| Discula destructiva virus 2 |
|
|
| Discula destructiva virus 2 – 331 | [AY033436, AY033437] | (DdV2-331) |
| Fusarium poae virus 1 |
|
|
| Fusarium poae virus 1 – A11 | [AF047013, AF015924] | (FpV1-A11) |
| Fusarium solani virus 1 |
|
|
| Fusarium solani virus 1 | [D55668, D55668] | (FsV1) |
| Gaeumannomyces graminis virus 019/6-A |
|
|
| Gaeumannomyces graminis virus 019/6-A |
| (GgV-019/6-A) |
| Gaeumannomyces graminis virus T1-A |
|
|
| Gaeumannomyces graminis virus T1-A |
| (GgV-T1-A) |
| Gremmeniella abietina RNA virus MS1 |
|
|
| Gremmeniella abietina RNA virus MS1 – C5 | [AY089993, AY089994, AY089995] | (GaRV-MS1-C5) |
| Helicobasidium mompa virus |
|
|
| Helicobasidium mompa virus | [AB025903] | (HmV) |
| Heterobasidion annosum virus |
|
|
| Heterobasidion annosum virus | [AF473549] | (HaV) |
| Ophiostoma partitivirus 1 |
|
|
| Ophiostoma partitivirus 1 | [AM087202, AM087203] | (OPV1) |
| Penicillium stoloniferum virus F |
|
|
| Penicillium stoloniferum virus F | [AY738336, AY738337] | (PsV-F) |
| Penicillium stoloniferum virus S |
|
|
| Penicillium stoloniferum virus S | [AY156521, AY156522] | (PsV-S) |
| Pleurotus ostreatus virus 1 |
|
|
| Pleurotus ostreatus virus 1 – South Korea | [AY533036, AY533038] | (PoV1-SKor) |
| Rhizoctonia solani virus 717 |
|
|
| Rhizoctonia solani virus 717 | [AF133290, AF133291] | (RhsV-717) |
| Rosellinia necatrix virus 1 |
|
|
| Rosellinia necatrix virus 1 – W8 | [AB113347, AB113348] | (RnV1-W8) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Partitivirus but have not been approved as species
| Botryotinia fuckeliana partitivirus 1 | [AM491609, AM491610, AM491611] | (BfPV1) |
| Ceratocystis polonica partitivirus 1 | [AY247204, AY247205] | (CpPV1) |
| Helicobasidium mompa partitivirus V1-1 | [AB110979] | (HmV-V1-1) |
| Helicobasidium mompa partitivirus V1-2 | [AB110980] | (HmV-V1-2) |
Genus Alphacryptovirus
Type species White clover cryptic virus 1
Distinguishing features
Members of the genus Alphacryptovirus infect plants and are transmitted from cell to cell during cell division. All members have two dsRNA segments that are believed to be individually encapsidated in separate particles.
Virion properties
Morphology
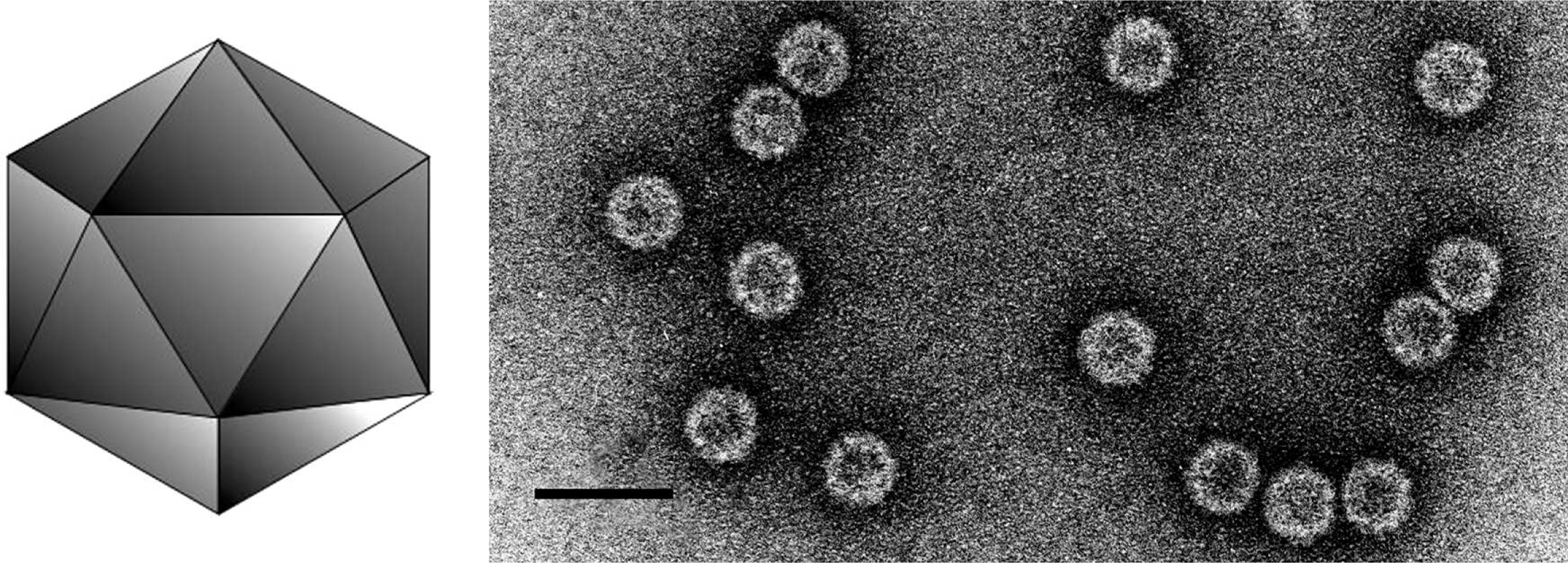
Virions are isometric, 30 nm in diameter. Particles lack fine structural detail, appearing rounded, usually penetrated by stain to give a ring-like appearance.
(Left) Diagrammatic representation of an alphacryptovirus capsid. (Right) Negative contrast electron micrograph of particles of an isolate of White clover cryptic virus 1, the type species of the genus Alphacryptovirus. The bar represents 50 nm.
(From Ghabrial et al., Family Partitiviridae. In: M.H.V. van Regenmortel et al. (Eds.), Virus Taxonomy: Seventh Report of the International Committee on Taxonomy of Viruses, Academic Press, San Diego, CA (2000), pp. 503-513; reproduced with permission).
Physicochemical and physical properties
Density in CsCl is 1.392 g cm−3.
Nucleic acid
The virions typically contain two dsRNA segments of about 1.5 and 2.0 kbp in size. The dsRNA segments are thought to be separately packaged.
Genome organization and replication
The genome typically consists of two dsRNA segments, whereas the larger one encodes the RdRp of about 73 kDa and the smaller one the CP of about 54 kDa. Conserved nucleotide sequence motifs of about seven to 12 nucleotides are located in the 5′-non translated region of dsRNA1 and dsRNA2 of beet cryptic virus 1 (BCV-1), Vicia cryptic virus (VCV), and white clover cryptic virus 1 (WCCV-1) as well as carrot cryptic virus (CaCV). In addition, these viruses contain conserved sequence motifs of 20 nucleotides in the 3′-non-translated region of dsRNA1 and 10 nucleotides in the 3′-non-translated region of dsRNA2, respectively. Both dsRNAs of the above mentioned viruses carry poly(A) stretches at their 3′ end, which may be interrupted by other nucleotides.
Antigenic properties
Some viruses in the genus are serologically related; none is related to viruses in the genus Betacryptovirus. There are no known serological relationships with fungal viruses in the genus Partitivirus.
Species demarcation criteria in the genus
The criteria to differentiate species in the genus are:
- Host range
- Size of dsRNA segments
- Serological relationships
Species in the genus are not serologically related (serological differentiation index of 5 or greater). Electrophoretic profiles of the genomic RNAs are distinct.
List of species in the genus Alphacryptovirus
| Alfalfa cryptic virus 1 |
|
|
| Alfalfa cryptic virus 1 |
| (ACV-1) |
| Beet cryptic virus 1 |
|
|
| Beet cryptic virus 1- Hungary | [EU489061, EU489062] | (BCV-1-Hun) |
| Beet cryptic virus 2 |
|
|
| Beet cryptic virus 2 |
| (BCV2) |
| Beet cryptic virus 3 |
|
|
| Beet cryptic virus 3 | [S63913] | (BCV3) |
| Carnation cryptic virus 1 |
|
|
| Carnation cryptic virus 1 |
| (CCV1) |
| Carrot temperate virus 1 |
|
|
| Carrot temperate virus 1 |
| (CteV1) |
| Carrot temperate virus 3 |
|
|
| Carrot temperate virus 3 |
| (CteV3) |
| Carrot temperate virus 4 |
|
|
| Carrot temperate virus 4 |
| (CteV4) |
| Hop trefoil cryptic virus 1 |
|
|
| Hop trefoil cryptic virus 1 |
| (HTCV1) |
| Hop trefoil cryptic virus 3 |
|
|
| Hop trefoil cryptic virus 3 |
| (HTCV3) |
| Radish yellow edge virus |
|
|
| Radish yellow edge virus |
| (RYEV) |
| Ryegrass cryptic virus |
|
|
| Ryegrass cryptic virus |
| (RGCV) |
| Spinach temperate virus |
|
|
| Spinach temperate virus |
| (SpTV) |
| Vicia cryptic virus |
|
|
| Vicia cryptic virus - Germany | [AY751737, AY75138] | (VCV-DE) |
| White clover cryptic virus 1 |
|
|
| White clover cryptic virus 1 | [AY705784, AY705785] | (WCCV1) |
| White clover cryptic virus 3 |
|
|
| White clover cryptic virus 3 |
| (WCCV3) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Alphacryptovirus but have not been approved as species
| Black raspberry cryptic virus | [EU082132] | (BrCV) |
| Carrot cryptic virus | [FJ550604, FJ550605] | (CaCV) |
| Cucumber cryptic virus |
| (CuCV) |
| Fragaria chiloensis cryptic virus | [DQ093961, DQ355440, DQ355439] | (FCCV) |
| Pepper cryptic virus 1 | [DQ361008] | (PCV1) |
| Pinus sylvestris cryptovirus | [AY973825] | (PSCV) |
| Pyrus pyrifolia cryptic virus | [AB012616] | (PpV) |
| Raphanus sativus cryptic virus 2 | [DQ218036, DQ218037, DQ218038] | (RsCV2) |
| Raphanus sativus cryptic virus 3 | [FJ461349, FJ461350] | (RsCV3) |
| Rose cryptic virus 1 | [EU413666, EU413667, EU413668] | (RoCV1) |
| (Rosa multiflora cryptic virus) |
|
|
Genus Betacryptovirus
Type species White clover cryptic virus 2
Distinguishing features
Members of the genus Betacryptovirus infect plants and are transmitted from cell to cell during cell division. All members have two dsRNA segments that are believed to be individually encapsidated in separate particles.
Virion properties
Morphology
Virions are isometric, 38 nm in diameter. Particles show prominent subunits, but their precise geometrical arrangement is not clear. The particles are rounded and are not penetrated by stain.
(Left) Diagrammatic representation of a betacryptovirus capsid. (Right) Negative contrast electron micrograph of particles of an isolate of White clover cryptic virus 2, the type species of the genus Betacryptovirus. The bar represents 50 nm.
(From Ghabrial et al., Family Partitiviridae. In: M.H.V. van Regenmortel et al. (Eds.), Virus Taxonomy: Seventh Report of the International Committee on Taxonomy of Viruses, Academic Press, San Diego, CA (2000), pp. 503-513; reproduced with permission).
Physicochemical and physical properties
Virion buoyant density in CsCl is 1.375 g cm−3.
Nucleic acid
Viral nucleic acid comprises two dsRNA segments, which are about 2.1 and 2.25 kbp.
Genome organization and replication
The genome typically consists of two dsRNA segments, whereas the larger one encodes the RdRp and the smaller one the putative CP.
Antigenic properties
Some viruses in the genus are serologically related; none is related to viruses in the genus Alphacryptovirus.
Species demarcation criteria in the genus
The criteria to differentiate species in the genus are:
- Host range
- Size of dsRNA segments
- Serological relationships
Species in the genus are not serologically related or are distantly related (serological differentiation index of 5 or greater). Electrophoretic profiles of the genomic RNAs are distinct.
List of species in the genus Betacryptovirus
| Carrot temperate virus 2 |
|
|
| Carrot temperate virus 2 |
| (CTeV2) |
| Hop trefoil cryptic virus 2 |
|
|
| Hop trefoil cryptic virus 2 |
| (HTCV2) |
| Red clover cryptic virus 2 |
|
|
| Red clover cryptic virus 2 |
| (RCCV2) |
| White clover cryptic virus 2 |
|
|
| White clover cryptic virus 2 |
| (WCCV2) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Betacryptovirus but have not been approved as species
| Alfalfa cryptic virus 2 |
| (ACV2) |
Genus Cryspovirus
Type species Cryptosporidium parvum virus 1
Distinguishing features
Members of the genus Cryspovirus infect apicomplexan protozoa of the genus Cryptosporidium and are largely transmitted by intracellular means during cell division and gamete fusion. Their genomes comprise two dsRNA segments of similar sizes that are individually encapsidated in separate particles.
Virion properties
Morphology
Virions are isometric and nonenveloped, about 31 nm in diameter as visualized by negative staining and transmission electron microscopy (Figure 7). The capsids appear single layered and thin, with short protrusions on their surfaces.
Physicochemical and physical properties
Virions have a buoyant density of 1.39–1.44 g cm−3 on CsCl gradients.
Nucleic acid
Virions contain two unrelated, linear dsRNA segments, 1.4 and 1.7 kbp in size, which are separately encapsidated. Additional dsRNA segments (satellite or defective) have not been reported.
Proteins
There is a single major CP sized 37 kDa and an RNA-dependent RNA polymerase sized 62 kDa. Virion-associated RNA polymerase activity is present.
Genome organization and replication
Each genome segment is monocistronic: dsRNA1 codes for the RdRp and dsRNA2 codes for the major CP. The virion-associated RdRp catalyzes in vitro end-to-end transcription of each dsRNA to produce mRNA by a semi-conservative mechanism.
Antigenic properties
Detection of the major CP by immunologic assays such as dot blotting has been reported as a means for identifying cryspovirus-positive Cryptosporidium isolates.
Biological properties
Infections of the Cryptosporidium host cells appear to be persistent and largely avirulent. Although Cryptosporidium species are pathogens of humans and other vertebrates, there are so far no well-established examples in which parasite pathogenicity is either positively or negatively modulated by cryspovirus infection. Virions are disseminated in nature within Cryptosporidium oocysts, which are shed profusely from Cryptosporidium-infected animals. A correlation between cryspovirus genome levels and parasite fecundity in terms of oocyst excretion has been reported.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Cryspovirus
| Cryptosporidium parvum virus 1 |
|
|
| Cryptosporidium parvum virus 1 – KSU-1 | [U95995, U95995] | (CSpV1-KSU1) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Cryspovirus but have not been approved as species
| Cryptosporidium felis virus | [DQ193520] | (CSfV) |
| Cryptosporidium hominis virus | [DQ193518] | (CShV) |
| Cryptosporidium meleagridis virus | [DQ193519] | (CSmV) |
List of related viruses which may be members of the family Partitiviridae but have not been approved as species
| Pyrus pyrifolia virus | [AB012616] | (PPV) |
Phylogenetic relationships within the family
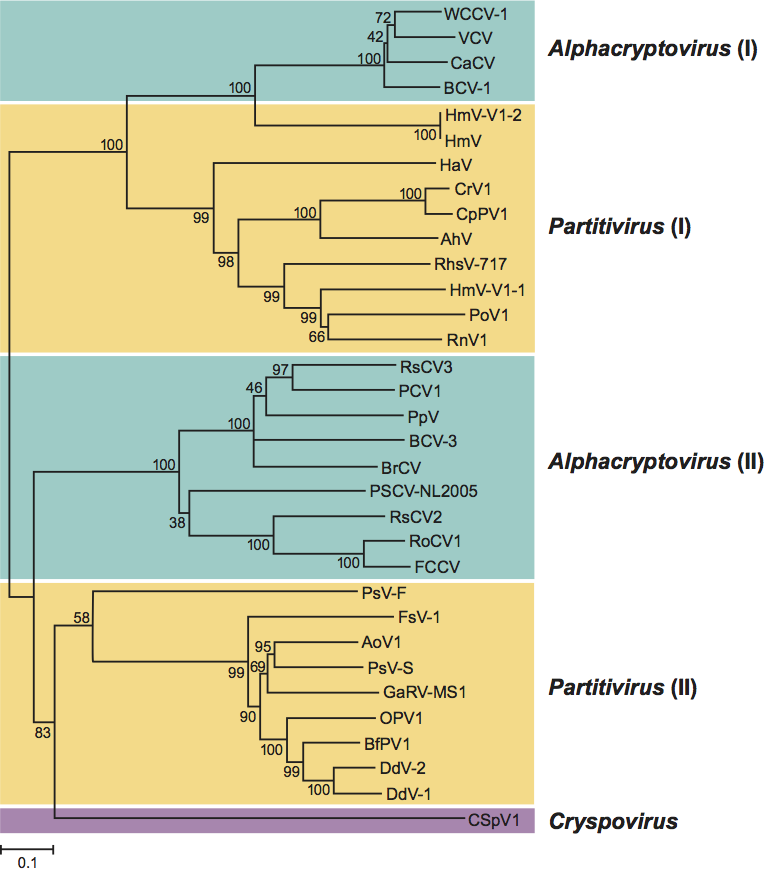
Phylogenetic analysis based on amino acid sequences of RdRps of members of the family Partitiviridae led to the identification of four clusters, two of which are large and comprise mostly members of the genus Partitivirus (Figure 8). One large cluster with strong bootstrap support includes an isolate of the type species AhV and related viruses (partitivirus I). The second cluster (partitivirus II) includes the well-characterized partitiviruses PsV-S and PsV-F. Members of cluster I have CPs and RdRps sized 70–77 and 77–87 kDa, respectively, whereas members of the partitivirus cluster II have CPs and RdRps sized 44–50 and 60–63 kDa, respectively. The genome segments of the two clusters are also differently sized, consistent with their different encoded protein sizes. Thus the genus Partitivirus can justifiably be divided into two new genera to reflect these two clusters. CSpV1, the only member of the genus Cryspovirus, is more closely related to viruses grouped into the partitivirus cluster II than to any other members of the family Partitiviridae. The remaining two RdRp clusters (Figure 8) comprise members of the genus Alphacryptovirus and probable alphacryptoviruses (see lists in the text above). One small cluster (alphacryptovirus I) consists of WCCV-1 (an isolate of the type species of the genus Alphacryptovirus) and two other approved alphacryptoviruses (BCV-1 and VCV) and a probable alphacryptovirus (CaCV). Interestingly, this small cluster of plant partitiviruses (cryptoviruses) is more closely related to the fungal partitiviruses in cluster partitivirus I than to the second cluster of alphacryptoviruses and probable alphacryptoviruses (alphacryptovirus II). The genus Alphacryptovirus can also thus justifiably be divided into two new genera to reflect these two clusters. No sequences have yet been reported for members of the genus Betacryptovirus, exempting them from this current analysis.
Similarity with other taxa
Members of the family Partitiviridae have properties similar to members of the family Picobirnaviridae, e.g., the genomes are bisegmented, and the capsids are small (<45 nm in diameter) with 120-subunit T=1 symmetry and share similar structural details. The picobirnaviruses, however, are phylogenetically distinct and infect vertebrates rather than plant, fungi and protozoa; they probably have other basic differences including co-packaging of both genome segments and capacity for extracellular transmission.
Derivation of names
Crypto: from Greek crypto, “hidden, covered, or secret”.
Cryspo: from the host genus name, Cryptosporidium.
Partiti: from Latin partitius, “divided”.
Further reading
Blawid, R., Stephan, D. and Maiss. E. (2008). Alphacryptovirus and Betacryptovirus. In: B.W.J. Mahy and M.H.V. Van Regenmortel (Eds.), Encyclopedia of Virology, 3rd edn, vol. 1. Elsevier, Oxford, pp. 68-75.
Boccardo, G., Milne, R.G., Luisoni, E., Lisa, V. and Accotto, G.P. (1985). Three seedborne cryptic viruses containing double-stranded RNA isolated from white clover. Virology, 147, 29-40.
Ghabrial, S.A., Ochao, W., Baker, T.B. and Nibert, M. (2008). Partitiviruses: General features. In: B.W.J. Mahy and M.H.V. Van Regenmortel (Eds.), Encyclopedia of Virology, 3rd edn, vol. 4. Elsevier, Oxford, pp. 68-75.
Khramtsov, N.V. and Upton, S.J. (2000). Association of RNA polymerase complexes of the parasitic protozoan Cryptosporidium parvum with virus-like particles: heterogeneous system. J. Virol., 74, 5788-5795.
Leoni, F., Gallimore, C.I., Green, J. and McLauchlin, J. (2006). Characterisation of small double stranded RNA molecule in Cryptosporidium hominis, Cryptosporidium felis and Cryptosporidium meleagridis. Parasitol. Int., 55, 299-306.
Nibert, M.L., Woods, K.M., Upton, S.J. and Ghabrial, S.A. (2009). Cryspovirus, a new genus of protozoan viruses in the family Partitiviridae. Arch. Virol., 154, 1959-1965.
Ochoa, W.F., Havens, W.M. Sinkovits, R.S., Nibert, M.L., Ghabrial, S.A. and Baker, T.S. (2008). Partitivirus structure reveals a 120-subunit, helix-rich capsid with distinctive surface arches formed by quasisymmetric coat-protein dimers. Structure, 16, 776-786.
Oh, C.-S. and Hillman, B.I. (1995). Genome organization of a partitivirus from the filamentous ascomycete Atkinsonella hypoxylon. J. Gen. Virol., 76, 1461-1470.
Pan, J., Dong, L., Lin, L., Ochoa, W.F., Sinkovits, R.S., Havens, W.M., Nibert, M.L., Baker, T.S., Ghabrial, S.A. and Tao, Y.J. (2009). Atomic structure reveals the unique capsid organization of a dsRNA virus. Proc. Natl. Acad. Sci., USA, 106, 4225-4230.
Sasaki, A., Kanematsu, S., Onoue, M., Oyama, Y. and Yoshida, K. (2006). Infection of Rosellinia necatrix with purified viral particles of a member of Partitiviridae (RnPV1-W8). Arch. Virol., 151, 697-707.
Szego, A., Enünlü, N., Deshmukh, S.D., Veliceasa, D., Hunyadi-Gulyás, E., Kühne, T., Ilyés, P., Potyondi, L., Medzihradszky, K. and Lukács, N. (2010). The genome of Beet cryptic virus 1 shows high homology to certain cryptoviruses present in phylogenetically distant hosts. Virus Genes, 40, 267-276.
Contributed by
Ghabrial, S.A., Nibert, M.L., Maiss, E., Lesker, T., Baker, T.S. and Tao, Y.J.
Figures
Figure 1 Transmission electron micrograph of negatively stained virions of an isolate of Penicilllium stoloniferum virus S, a representative species of the genus Partitivirus.
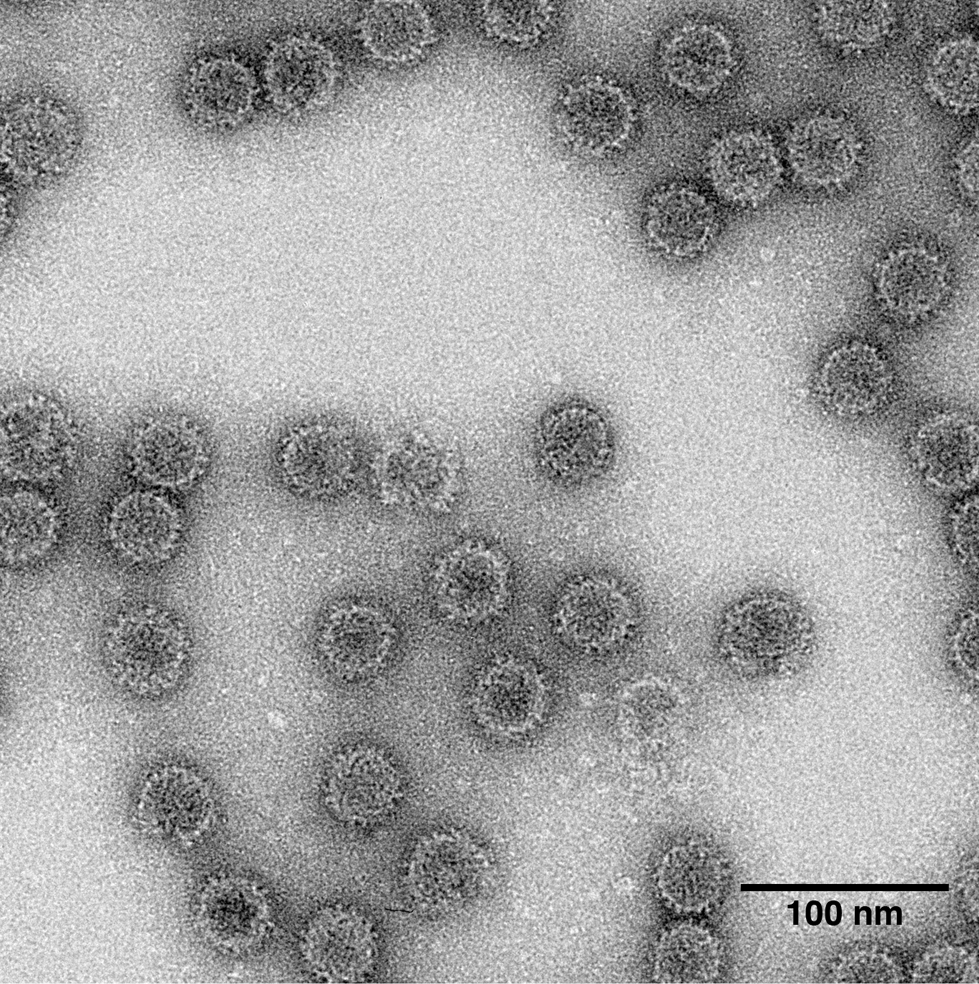
Figure 2 Structures of two partitiviruses. (A) Crystal structure of an isolate of Penicillium stoloniferum virus F. Two asymmetric units of the icosahedron are highlighted along with the icosahedral symmetry elements that define the boundaries of these units. One CPA monomer (in red) and one CPB monomer (in yellow) from the same CP dimer are labelled. (B-C) Cryo-EM reconstructions of PsV-F and -S, both at ~4.5 resolution, and rendered with radial, color mapping. (D) Putative capsid assembly unit consists of a dimer of CP dimers (redyellow, labelled A1-B1 and pinkorange, labelled A2-B2) that occupy a diamond-shaped area defined by black lines. (E) A PsV-F CP dimer. In this and the next panel (F), the arch and shell domains are highlighted green/light green and red/magenta, respectively, and the two subunits are distinguished as green or light green and red or magenta. (F) A PsV-S CP dimer. The atomic coordinates for this model were generated from a homology model derived from the PsV-F structure and constrained by its fit to the PsV-S cryo-EM reconstruction.
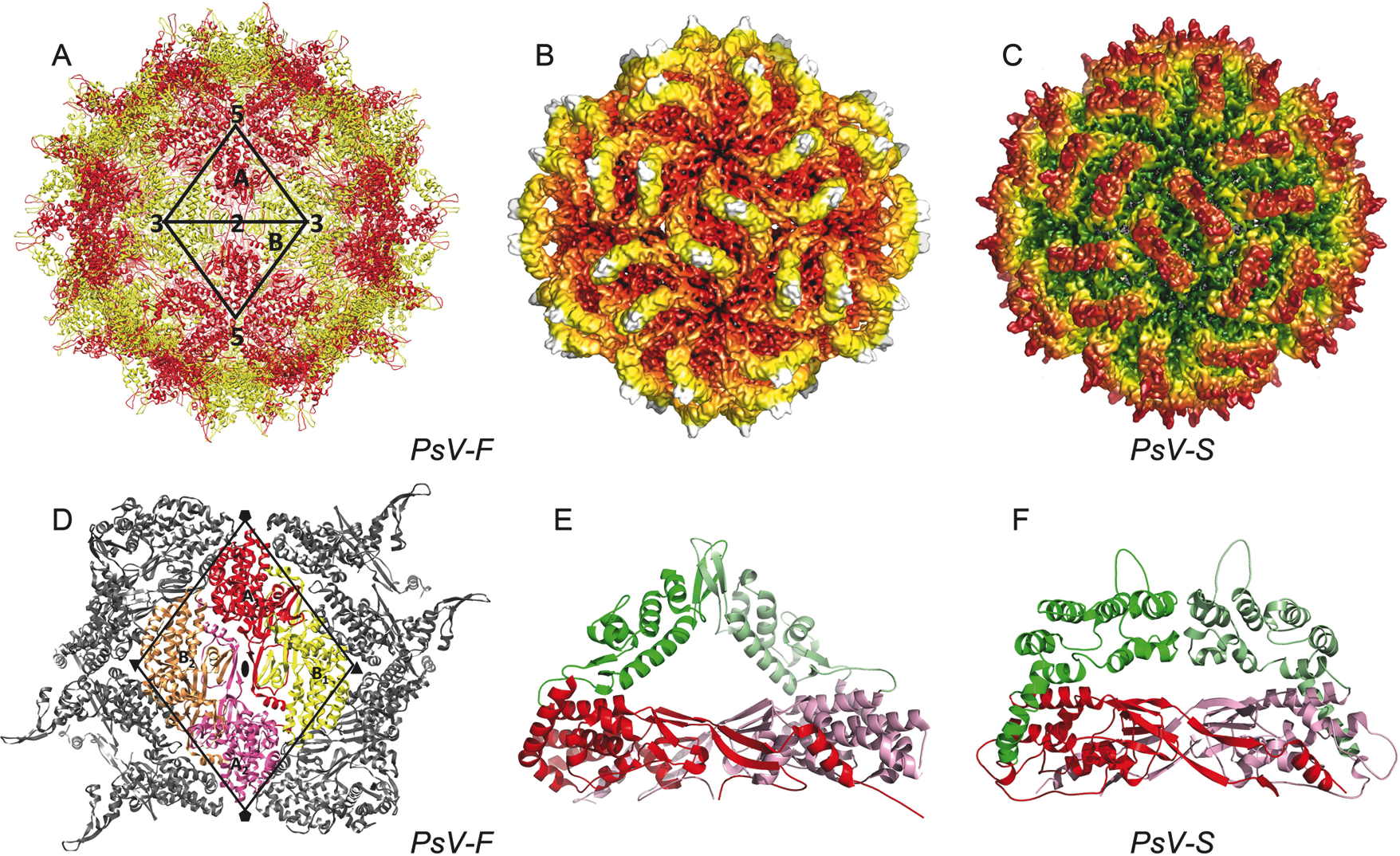
Figure 3 Genome organization of Atkinsonella hypxylon virus (AhV). The RdRp ORF (nt positions 40-2038 on dsRNA1) and the CP ORF (nt positions 72-2030 on dsRNA2) are represented by rectangular boxes.
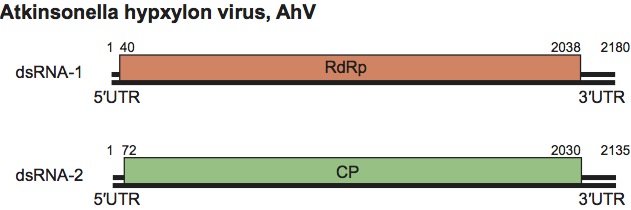
Figure 4 Model for replication of Penicillium stoloniferum virus S (PsV-S). The open circles represent CP subunits and the closed circles represent RNA polymerase subunits. Solid lines represent RNA strands whereas wavy lines represent mRNA.
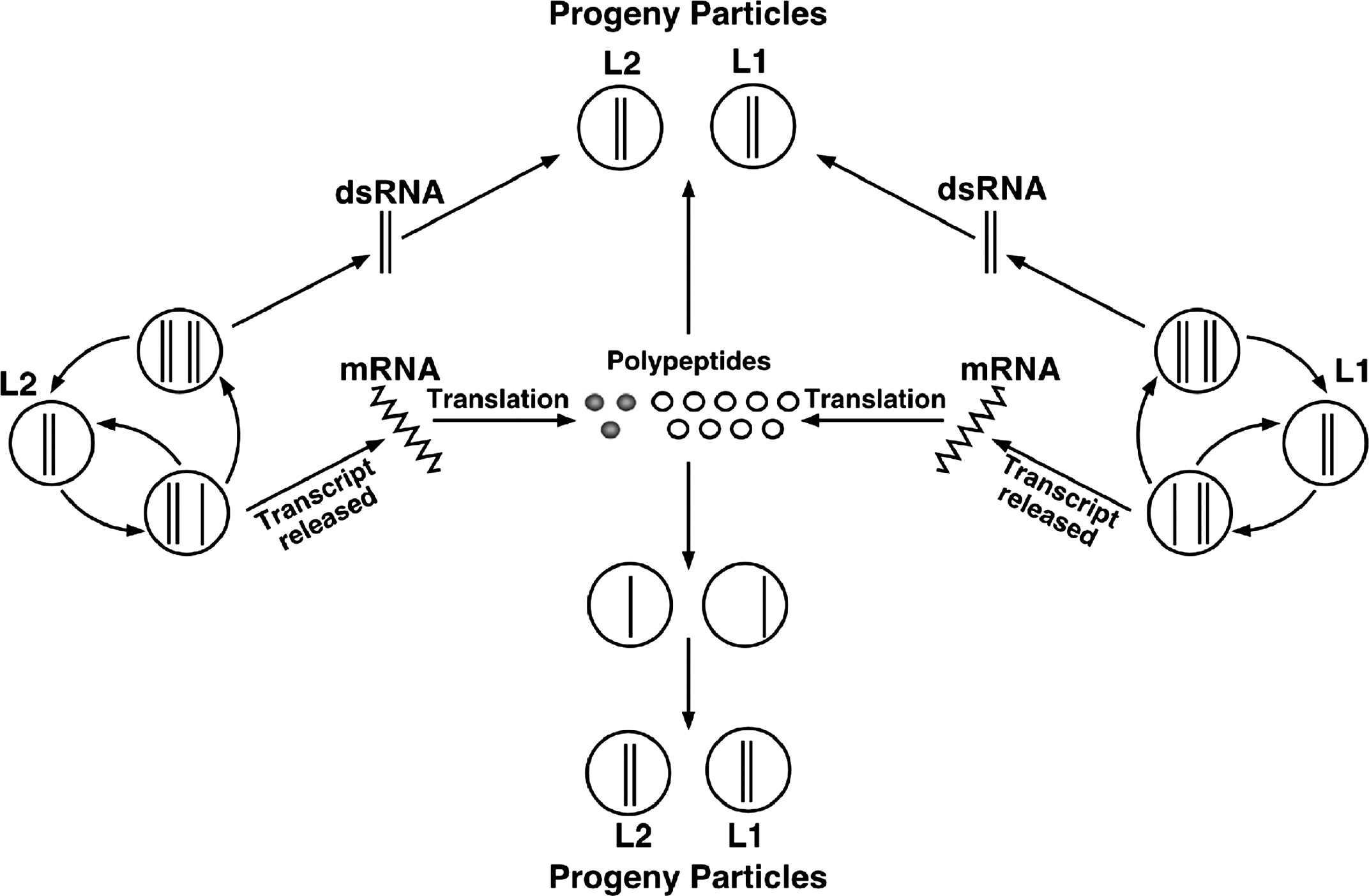
Figure 5 (Left) Diagrammatic representation of an alphacryptovirus capsid. (Right) Negative contrast electron micrograph of particles of an isolate of White clover cryptic virus 1, the type species of the genus Alphacryptovirus. The bar represents 50 nm.
(From Ghabrial et al., Family Partitiviridae. In: M.H.V. van Regenmortel et al. (Eds.), Virus Taxonomy: Seventh Report of the International Committee on Taxonomy of Viruses, Academic Press, San Diego, CA (2000), pp. 503-513; reproduced with permission).
Figure 6 (Left) Diagrammatic representation of a betacryptovirus capsid. (Right) Negative contrast electron micrograph of particles of an isolate of White clover cryptic virus 2, the type species of the genus Betacryptovirus. The bar represents 50 nm.
(From Ghabrial et al., Family Partitiviridae. In: M.H.V. van Regenmortel et al. (Eds.), Virus Taxonomy: Seventh Report of the International Committee on Taxonomy of Viruses, Academic Press, San Diego, CA (2000), pp. 503-513; reproduced with permission).
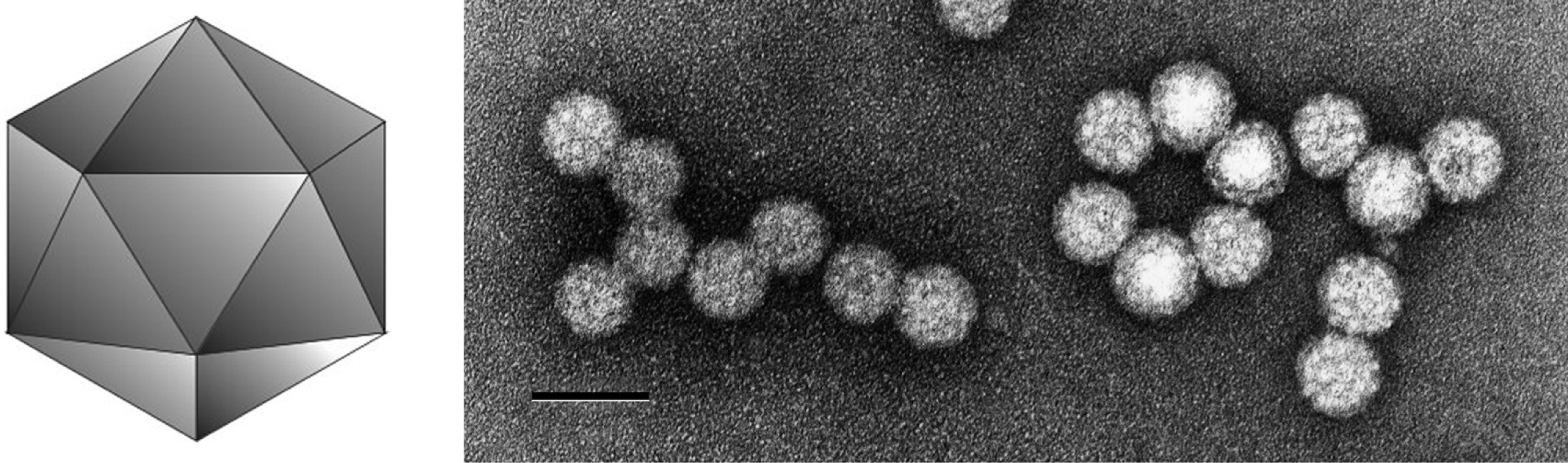
Figure 7 Electron micrograph of Cryptosporidium parvum virus 1 particles. Particles were gradient-purified from C. parvum strain KSU-1, negatively stained with uranyl acetate and visualized by transmission electron microscopy. Bar represents 100nm.
(Image is reproduced from Khramtsov and Upton (2000) with permission of the American Society for Microbiology.)
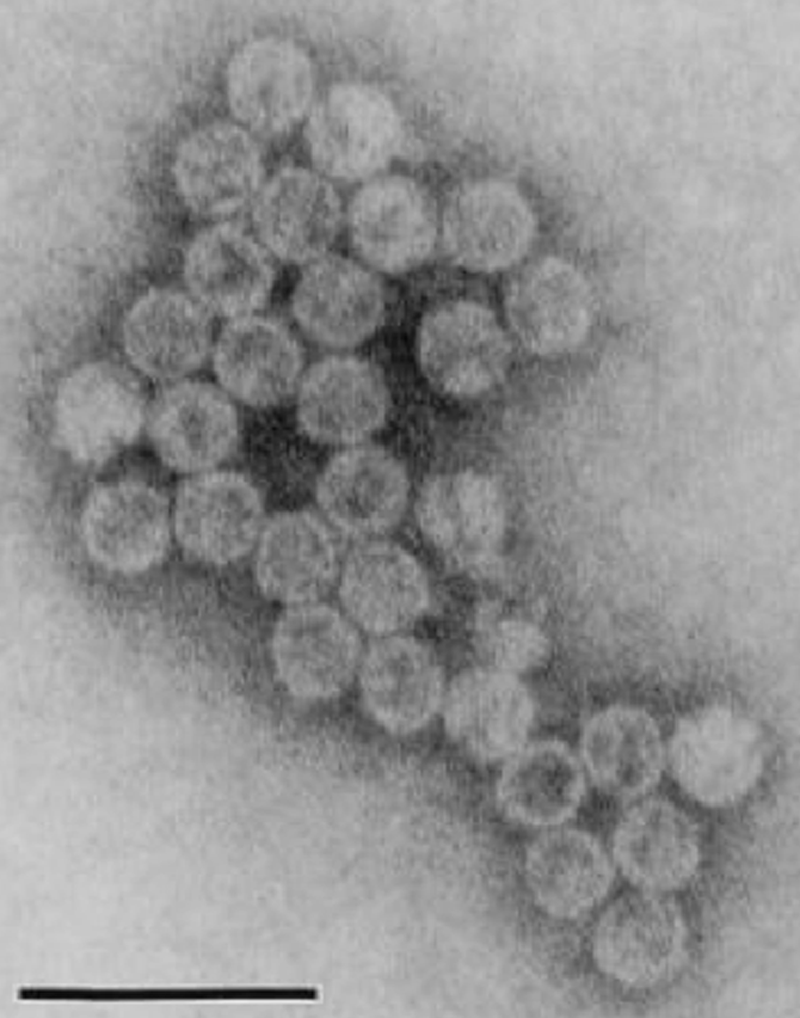
Figure 8 Neighbor-joining phylogenetic tree constructed based on the complete amino acid sequences of RdRps of members and probable members of the family Partitiviridae. The amino acid sequences were aligned using the program CLUSTAL W. For virus names and abbreviations, see lists in text. The phylogenetic tree was generated including codon positions using the MEGA4 phylogenetic package. Bootstrap percentages out of 1000 replicates are indicated at the nodes.