Family: Endornaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Endornavirus
Type species: Vicia faba endornavirus
Virion properties
Morphology
None reported. Endornaviruses do not produce virions.
Physicochemical and physical properties
None reported.
Nucleic acid
The linear dsRNA genomes of these viruses range in length from about 14 kbp to about 17.6 kbp. Each characterized genome includes a site-specific break (nick) in the coding strand about 1.2–2.7 kbp from the 5′ terminus.
Proteins
None yet characterized. RdRp activity has been detected in cytoplasmic vesicles, which also contain the genomic dsRNA. Endornaviruses lack virion proteins.
Lipids
None yet characterized. A lipid membrane that is probably derived from the host bounds the cytoplasmic vesicles.
Carbohydrates
None yet characterized. Carbohydrate, possibly a glycolipid, has been detected in preparations of the cytoplasmic vesicles in plants infected with endornaviruses.
Genome organization and replication
Each characterized genome encodes a single long polypeptide that crosses the break in the coding strand. These polypeptides include aa sequences typical of viral RNA helicases (Hels), UDP-glucosyltransferases (UGTs) and RNA-dependent RNA polymerases (RdRps). The polypeptides of Oryza sativa endornavirus (OsEV), Oryza rufipogon endornavirus (OrEV) and Phytophthora endornavirus 1 (PEV1) are about 4600 aa residues long, and those of Helicobasidium mompa endornavirus 1 (HmEV1) and Vicia faba endornavirus (VfEV) are about 5500 aa residues long. RNA replication occurs in cytoplasmic vesicles where RdRp activity has been detected in association with the genomic dsRNA. The cytoplasmic vesicles, sometimes called “virus-like particles,” are bounded by a unit membrane and are believed to be functionally equivalent to the replication complexes of positive strand RNA viruses. Endornavirus RNA has been found in every tissue and at every developmental stage and is maintained at an almost constant concentration (20–100 copies/cell) except in the pollen of some species.
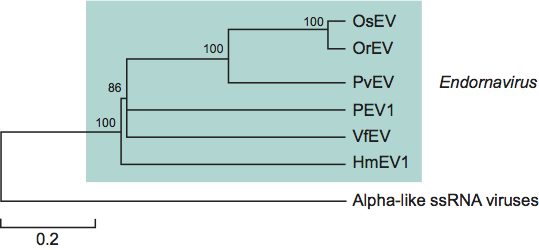
A genome map for an isolate of Oryza sativa endornavirus. A triangle marks the position of the break in the coding strand (1221 nucleotide from the 5′ end of the coding strand). Hel, UGT and RdRp indicate the positions of viral RNA helicase, UDP-glucosyltransferase and RNA-dependent RNA polymerase domains, respectively.
Antigenic properties
Monoclonal antibodies raised against purified cytoplasmic vesicles from plants infected with VfEV allowed the VfEV-associated male sterility in the progeny of crosses to be detected. The antibodies recognized an epitope that contains sugars, possibly a glycolipid.
Biological properties
Endornaviruses are found in some plants, fungi and oomycetes. Natural infections of endornaviruses have been confirmed in some varieties of cultivated rice (Oryza sativa), wild rice (Oryza rufipogon), broad bean (Vicia faba) and kidney bean (Phaselous vulgaris). Other plants that may be infected by species from this family include barley, bottle gourd, Malabar spinach, melon and pepper. Plant endornaviruses are transmitted through seed via both ova and pollen. No horizontal spread has been observed in the field and no potential vectors have been identified. No attempt to transmit the viruses other than through seed has succeeded, although the deduced phylogeny suggests that inter-species transmission has occurred in the past. Plant endornaviruses are not mechanically transmissible. None is associated with disease symptoms except for VfEV, which induces cytoplasmic male sterility.
A large dsRNA in the V670 strain of Helicobasidium mompa, the violet root rot fungus, has been identified as a hypovirulence factor, and shown by sequencing to be an endornavirus (Helicobasidium mompa endornavirus 1). Similarly, a 13.9 kbp dsRNA isolated from a Phytophthora sp. from Douglas fir (Pseudotsuga sp.) was identified as a member of the genus (Phytophthora endornavirus 1) by sequencing.
Species demarcation criteria in the genus
At present, species are distinguished on the basis of their host-range and sequence differences. Each recognized endornavirus species was isolated from a different host species. The genomic nucleotide sequences of different endornavirus species are only 30% to 75% identical.
List of species in the genus Endornavirus
| Infecting plants |
|
|
| Oryza sativa endornavirus |
|
|
| Oryza sativa endornavirus - Nipponbare | [D32136] | (OsEV-Nip) |
| Oryza rufipogon endornavirus |
|
|
| Oryza rufipogon endornavirus W-1714 | [AB014344] | (OrEV-W-1714) |
| Vicia faba endornavirus |
|
|
| Vicia faba endornavirus - 447 | [AJ000929] | (VfEV-447) |
| Phaseolus vulgaris endornavirus |
|
|
| Phaseolus vulgaris endornavirus - Black Turtle Soup | [AB185246] | (PvEV-BTS) |
| Infecting fungi |
|
|
| Helicobasidium mompa endornavirus 1 |
|
|
| Helicobasidium mompa endornavirus 1-670 | [AB218287] | (HmEV1-670) |
| Infecting oomycetes |
|
|
| Phytophthora endornavirus 1 |
|
|
| Phytophthora endornavirus 1 - Oregon | [AJ877914] | (PEV1-OR) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Endornavirus but have not been approved as species
| Infecting fungi |
|
|
| Gremmeniella abietina type B RNA virus XL | [DQ399289] | (GaBRV-XL) |
Phylogenetic relationships within the family
Phylogenetic relationships within the family are depicted in Figure 2.
Similarity with other taxa
Comparisons and analyses of RdRp and helicase sequences indicate that endornaviruses are related to viruses of the “alpha-like” supergroup.
Derivation of name
Endorna: from endo, Greek “within”, and RNA.
Further reading
Fukuhara, T. and Moriyama, H. (2008). Endornaviruses. In: B.W.J. Mahy and M.H.V. van Regenmortel (Eds.), Encyclopedia of Virology, 3rd edn. Elsevier, Oxford, pp. 109–116.
Fukuhara, T., Moriyama, H., Pak, J.K., Hyakutake, T. and Nitta, T. (1993). Enigmatic double-stranded RNA in Japonica rice. Plant Mol. Biol., 21, 1121–1130.
Gibbs, M.J., Koga, R., Moriyama, H., Pfeiffer, P. and Fukuhara, T. (2000). Phylogenetic analysis of some large double-stranded RNA replicons from plants suggests they evolved from a defective single-stranded RNA virus. J. Gen. Virol., 81, 227–233.
Hacker, C.V., Brasier, C.M. and Buck, K.W. (2005). A double-stranded RNA from a Phytophthora species is related to the plant endornaviruses and contains a putative UDP glycosyltransferase gene. J. Gen. Virol., 86, 1561–1570.
Lefebvre, A., Scalla, R. and Pfeiffer, P. (1990). The double-stranded RNA associated with the '447' cytoplasmic male sterility in Vicia faba is packaged together with its replicase in cytoplasmic membranous vesicles. Plant Mol. Biol., 14, 477–490.
Moriyama, H., Horiuchi, H., Koga, R. and Fukuhara, T. (1999). Molecular characterization of two endogenous double-stranded RNAs in rice and their inheritance by interspecific hybrids. J. Biol. Chem., 274, 6882–6888.
Osaki, H., Nakamura, H., Sasaki, A., Matsumoto, N. and Yoshida, K. (2006). An endornavirus from a hypovirulent strain of the violet root rot fungus, Helicobasidium mompa. Virus Res., 118, 143–149.
Pfeiffer, P. (1998). Nucleotide sequence, genetic organization and expression strategy of the double-stranded RNA associated with the '447' cytoplasmic male sterility trait in Vicia faba. J. Gen. Virol., 79, 2349–2358.
Pfeiffer, P. (2002). Large dsRNA genetic elements in plants and the novel dsRNA associated with the "447" cytoplasmic male sterility in Vicia faba. In: S.M. Tavantzis (Ed.), dsRNA Genetic Elements: Concepts and Applications in Agriculture, Forestry, and Medicine. CRC Press, Boca Raton, FL, pp. 259–274.
Contributed by
Fukuhara, T. and Gibbs, M.J.
Figures
Figure 1 A genome map for an isolate of Oryza sativa endornavirus. A triangle marks the position of the break in the coding strand (1221 nucleotide from the 5 end of the coding strand). Hel, UGT and RdRp indicate the positions of viral RNA helicase, UDP-glucosyltransferase and RNA-dependent RNA polymerase domains, respectively.
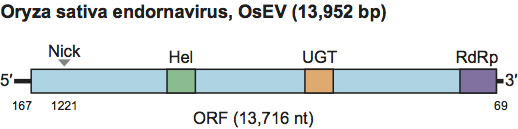
Figure 2 Phylogenetic positions of endornaviruses. About 470 aa of the RdRp regions of six endornaviruses and 10 alpha-like ssRNA viruses were analyzed by using the ClustalX and MEGA2 (Molecular Evolutionary Genetics Analysis) programs, and the resulting neighbor-joining (NJ) tree is shown. A bootstrap test was performed with 100 resamplings.