Family: Cystoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Cystovirus
Type species Pseudomonas phage phi6
Distinguishing features
The virion is enveloped and contains a segmented dsRNA genome. The innermost protein capsid is a polymerase complex responsible for genome packaging, replication and transcription.
Virion properties
Morphology
The enveloped virions are spherical, about 85 nm in diameter and covered by spikes (Figure 1). The envelope surrounds an isometric nucleocapsid, about 58 nm in diameter. The nucleocapsid surface shell (if present) follows T=13 isocahedral symmetry. Turret-like extrusions of the underlying polymerase complex span the nucleocapsid surface shell layer at the five-fold symmetry positions (Figure 1). The dodecahedral polymerase complex is about 50 nm in diameter. In the polymerase complex major capsid protein dimers are arranged on a T=1 icosahedral lattice.
Physicochemical and physical properties
The molecular mass of Pseudomonas phage phi6 virion is 99 ×106 and nucleocapsid 40 ×106. Virion S20,w is about 405S. The buoyant density of the virion is 1.27 g cm−3 in CsCl, 1.22 g cm−3 in Cs2SO4 and 1.24 g cm−3 in sucrose. Virions are sensitive to detergents, ether and chloroform but stable between pH 6.0 and 9.5.
Nucleic acid
Virions contain three segments of linear, double stranded RNA: L (6.4–7.1 kb), M (3.6–4.7 kb), and S (2.6–3.2 kb). The complete genome is 12.7–15.0 kb and has a guanine+cytosine content of approximately 56%. All the genome segments are enclosed in a single particle and each virion contains a single copy of the genome. The genome constitutes approximately 10% of the virion weight.
Proteins
Proteins constitute about 70% of the virion weight. The viral genome (Figure 2) encodes structural (Figure 1) and non-structural proteins.
The envelope contains three integral membrane proteins: P6, P9 and P10. Additional membrane proteins with unknown function are found from some members of the genus. Receptor binding spike is anchored to the envelope via fusogenic protein P6. The spike complex is composed of one (P3) to three polypeptides (P3a, P3b, P3c). Protein P8 forms the nucleocapsid surface shell (Figure 1) or is part of the envelope. Protein P5 is a lytic enzyme associated on the nucleocapsid surface. Major capsid protein P1 (120 copies per virion) is involved in single stranded RNA binding. Protein P2 is the viral replicase and transcriptase and is located in the interior of the P1 shell. The turret-like extrusions of the polymerase complex are made by hexamers of P4 protein. P4 is a nucleoside triphosphatase required for genome packaging and transcription. Minor capsid protein P7 is an assembly factor.
Non-structural protein P12 is a morphogenetic protein that is involved in envelope assembly. Non-structural proteins J and Hb of Pseudomonas phage phi8 regulate the stability of viral message RNAs. Different members of the genus may also encode additional non-structural proteins with unknown function.
Lipids
Virions are composed of 20% lipids by weight. There is enough lipid to cover about one-half of the envelope surface area (the rest being protein). Viral lipids are derived from host plasma membrane. The lipid compositions of viral envelope and host plasma membrane are similar.
Carbohydrates
None reported.
Genome organization and replication
The overall genetic organization is similar in all proposed members of the genus. Genes are clustered into functional groups (Figure 2). Distinct noncoding regions at the termini of the three genome segments contain signals for genome packaging and replication.
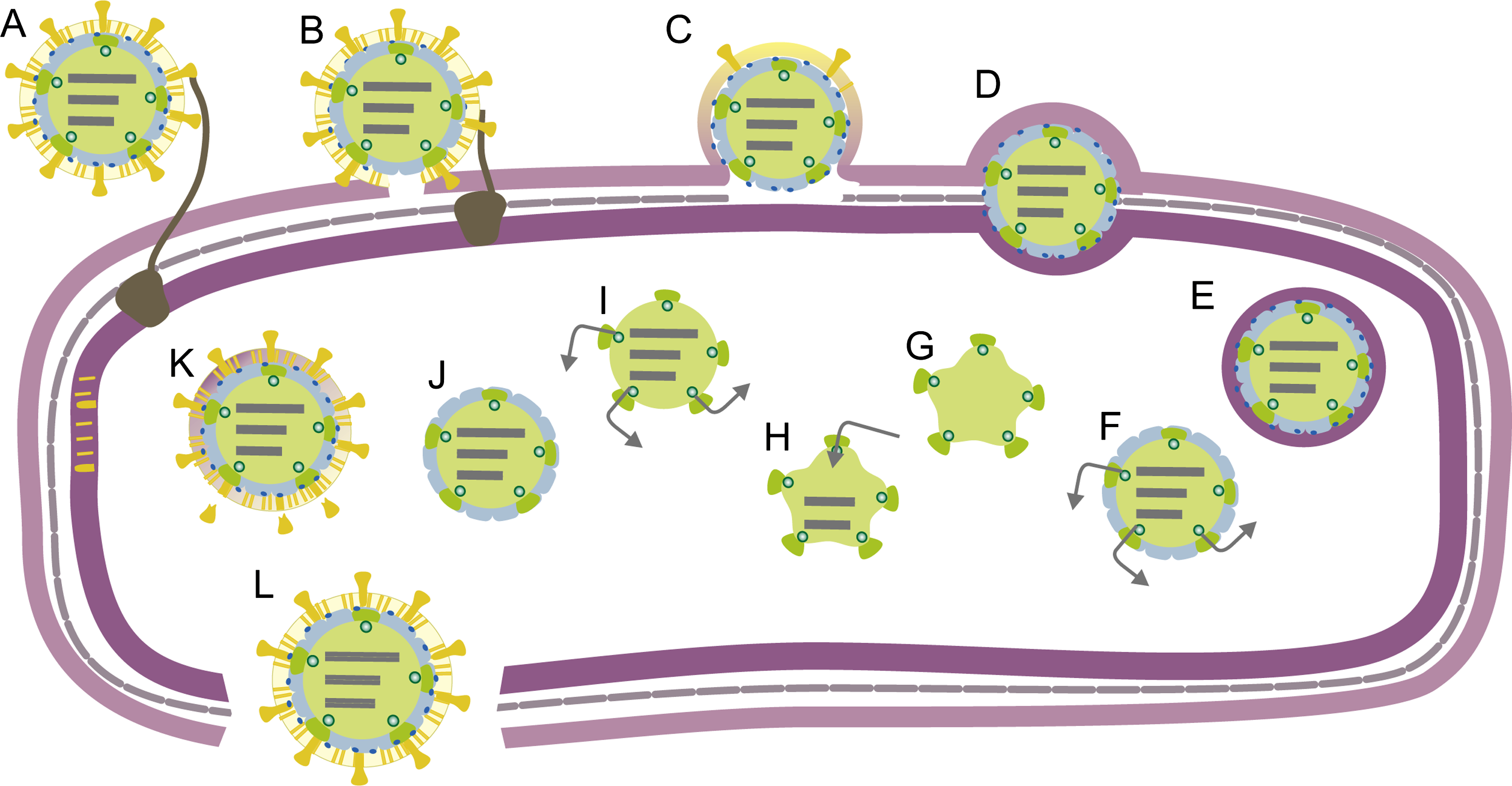
Virions adsorb to pili or in some viruses directly to the outer membrane of the host bacterium (Figure 3). Viral envelope fuses with the host outer membrane and the nucleocapsid associated lytic enzyme locally digests the peptidoglycan layer. Viral polymerase complex delivery into the host cytoplasm involves an endocytic-like process at the host plasma membrane. The viral genome is transcribed by virion-associated RNA-dependent RNA polymerase within the polymerase complex. Early in the infection approximately equal amounts of messenger RNA molecules are produced from L, M and S. Later in the infection cycle transcripts of M and S typically predominate. This temporal control relies either on host factors or viral nonstructural proteins. Transcription is semi-conservative and produces full-length copies of the genome segments.
The produced messenger RNA molecules are polycistronic. Translation of L transcripts produces the early proteins, which assemble to form empty polymerase complexes (Figure 3). These package the three positive strand transcripts. Negative strand synthesis then takes place inside the polymerase complex. RNA packaging and replication induce structural changes in the polymerase complex (expansion). Transcription by these polymerase complexes produces messages for late protein synthesis. The nucleocapsid surface shell (if present) assembles on the polymerase complex (Figure 3) and inactivates transcription. Nucleocapsid acquires protein P5 and the envelope. Spikes are assembled on the envelope surface. Mature virions are release upon virus-induced host cell lysis.
Antigenic properties
No information available.
Biological properties
Cystoviruses are lytic bacteriophages that induce host cell lysis at the end of the viral reproduction cycle. Natural hosts are gram negative plant pathogenic bacteria.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Cystovirus
| Pseudomonas phage phi6 |
|
|
| Pseudomonas phage phi6 | [L: M17461; M: M17462; S: M12921] | (phi6) |
Species names are in italic script; names of strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Cystovirus but have not been approved as species
| Pseudomonas phage phi7 |
| (phi7) |
| Pseudomonas phage phi8 | [L:AF226851; M:AF226852; S:AF226853] | (phi8) |
| Pseudomonas phage phi9 |
| (phi9) |
| Pseudomonas phage phi10 |
| (phi10) |
| Pseudomonas phage phi11 |
| (phi11) |
| Pseudomonas phage phi12 | [L:AF408636; M:AY039807; S:AY034425] | (phi12) |
| Pseudomonas phage phi13 | [L:AF261668; M:AF261667; S:AF261666] | (phi13) |
| Pseudomonas phage phi14 |
| (phi14) |
| Pseudomonas phage phi2954 | [L:FJ608823; M:FJ608824; S:FJ608825] | (phi2954) |
Phylogenetic relationships within the family
Not applicable.
Similarity with other taxa
In terms of genome replication strategy, cystoviruses resemble eukaryotic double stranded RNA viruses. The structure, organization and functions of the polymerase complex containing the genome are the major similarities among members of families Cystoviridae, Reoviridae, Totiviridae, Partitiviridae and Picobirnaviridae. The T=13 architecture of the surrounding capsid layer is also shared by cystoviruses and reoviruses.
Derivation of name
Cysto: from Greek kystis, “bladder”, “sack”.
Further reading
Ackermann and DuBow, 1987 H.-W. Ackermann, M.S. DuBow, H.-W. Ackermann, M.S. DuBow, CystoviridaeViruses of Prokaryotes, Volume II: Natural Groups of Bacteriophages. In: H.-W. Ackermann, M.S. DuBow, Viruses of Prokaryotes, Volume II: Natural Groups of Bacteriophages. CRC Press, Boca Raton, FL1987171–218.
Bamford et al., 2005 D.H. Bamford, J.M. Grimes, D.I. Stuart, What does structure tell us about virus evolution?. Curr. Opin. Struct. Biol. 15 (2005) 655–663.
Mertens, 2004 P. Mertens, The dsRNA viruses. Virus Res. 101 (2004) 3–13.
Mindich, 2006 L. Mindich, R. Calendar, Phages with segmented double-stranded RNA genomesThe Bacteriophages. In: R. Calendar, The Bacteriophages. Oxford University Press, New York2006197–207.
Mindich et al., 1999 L. Mindich, X. Qiao, J. Qiao, S. Onodera, M. Romantschuk, D. Hoogstraten, Isolation of additional bacteriophages with genomes of segmented double-stranded RNA. J. Bacteriol. 181 (1999) 4505–4508.
Poranen et al., 2005 M.M. Poranen, R. Tuma, D.H. Bamford, Assembly of double-stranded RNA bacteriophages. Adv. Virus Res. 64 (2005) 15–43.
Qiao et al., 2010 X. Qiao, Y. Sun, J. Qiao, F. Di Sanzo, L. Mindich, Characterization of φ2954, a newly isolated bacteriophage containing three dsRNA genomic segments. BMC Microbiol. 10 (2010) 1471–2180.
Contributed by
Poranen, M.M. and Bamford, D.H.
Figures
Figure 1 (Left) Schematic presentation of cystovirus particle (Pseudomonas phage phi6) with location of virion proteins. (Middle) Three-dimensional reconstructions of the nucleocapsid. (Right) Thin-section electron micrograph of Pseudomonas phage phi6 particles attached to the pilus receptor. The bar represents 200 nm.
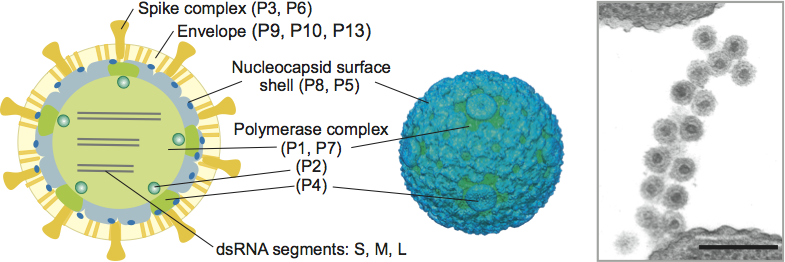
Figure 2 Genome organization of Pseudomonas phage phi6. The gene and protein numbers are the same. Genes encoding constituents of the polymerase complex and nucleocapsid are in green and blue, respectively. Genes encoding envelope associated proteins are in purple, and non-structural proteins in orange.
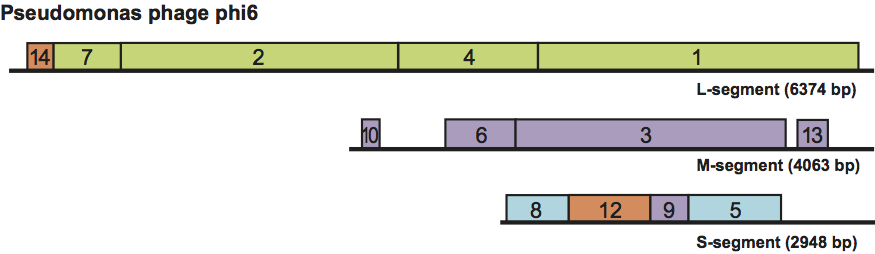
Figure 3 Schematic of the Pseudomonas phage phi6 life cycle. (A) Adsorption. (B) Envelope fusion. (C) Peptidoglycan digestion. (DE) Endocytotic uptake of nucleocapsid. (F) Early transcription. (G) Polymerase complex assembly. (H) ssRNA packaging and replication. (I) Late transcription. (J) Nucleocapsid shell assembly. (K) Translocation of the viral envelope and assembly of spikes. (L) Host cell lysis and release of mature virions.