Family: Chrysoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Chrysovirus
Type species Penicillium chrysogenum virus
Virion properties
Morphology
Virions are isometric, non-enveloped, about 400 Å in diameter, and the protein shell is 48 Å thick. The capsid comprises 60 protein subunit monomers arranged on a T=1 icosahedral lattice; the most prominent features are 12 outward-protruding pentamers. The capsid protein has a high content of α helices, forming a repeated α-helical core indicative of gene duplication (or joined folds). Whereas the Penicillium chrysogenum virus (PcV) capsid protein has two motifs with the same fold, most dsRNA virus capsid subunits consist of dimers of a single protein with similar folds. The spatial arrangement of the α-helical core resembles that found in the L-A virus capsid protein, a fungal totivirus, suggesting a conserved basic fold. Due to the numerous interactions of the encapsidated genome with the inner surface of the capsid, specifically six interacting areas per monomer, the outermost dsRNA layer is arranged in an icosahedral cage, and is partially visualized as tubular densities corresponding to an A-form duplex. The genome ordering might constitute a framework for dsRNA transcription at the capsid interior, and/or has a structural role for capsid stability.
Physicochemical and physical properties
Virion buoyant density in CsCl is in the range of 1.34–1.39 g cm−3and S20,w is 145-150S.
(Left) Electron micrographs of Penicillium chrysogenum virus (PcV), the type species of the genus Chrysovirus. Samples were negatively stained in 2% uranyl acetate (lower panel) or prepared unstained and vitrified (upper panel); the bar represents 50 nm. (Right) Three-dimensional cryo-electron microscopy reconstruction of PcV virions at 8 Å resolution. Surface-shaded virion capsid viewed along an icosahedral twofold axis. Boundaries for two capsid proteins are outlined in black; each subunit has two similar domains (red and green), suggesting ancestral gene duplication. Icosahedral symmetry axes are indicated.
Nucleic acid
Virions contain four unrelated linear, separately encapsidated, dsRNA segments (2.4–3.6 kbp in size, Table 1). The largest segment, dsRNA-1, codes for the virion-associated RNA polymerase and dsRNA-2 codes for the major CP. Both dsRNAs 3 and 4 encode proteins of unknown function. Sequences at the 5′- and 3′-UTRs are highly conserved among the four-dsRNA segments. The 5′-UTRs are relatively long, between 140 and 400 nt (nucleotides) in length. In addition to the absolutely conserved 5′- and 3′-termini, a 40–75 nt region with high sequence identity is present in the 5′-UTR of all four dsRNAs (Box 1; Figure 2). A second region of strong sequence similarity is present immediately downstream from Box 1 (Figure 2). This consists of a stretch of 30–50 nt containing a reiteration of the sequence “CAA”. The (CAA)n repeats are similar to the enhancer elements present at the 5′-UTRs of tobamoviruses.
Table 1 List of the dsRNA segments of Penicillium chrysogenum virus (PcV), with their size (bp), calculated size of their encoded proteins and function
| Segment | Size (bp)a | Size of encoded protein (Da)b | Function of protein |
| dsRNA1 | 3562 | 128,548 (1117 aa) | RdRp |
| dsRNA2 | 3200 | 108,806 (982 aa) | CP |
| dsRNA3 | 2976 | 101,458 (912 aa) | Unknown |
| dsRNA4 | 2902 | 94,900 (847 aa) | Unknown |
a Size determined by sequencing full-length cDNA clones of the indicated genome segments.
b Size determined from the predicted aa sequences derived from full-length cDNA clones of the indicated genome segments; The number of aa residues in the encoded viral proteins are placed in parentheses.
Proteins
The capsids are made up of a single major polypeptide species (110–112 kDa). Virion-associated RNA polymerase activity is present.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
PcV has a multipartite genome comprising four unrelated linear dsRNA segments (Figure 2). Each is monocistronic; dsRNA-1 codes for the RdRp and dsRNA2 codes for the major CP. Although the proteins P3 and P4, coded for by dsRNA-3 and dsRNA-4, respectively, are of unknown function, protein database searches reveal that PcV P3 sequence shares a “phytoreo S7 domain” with a family consisting of several phytoreovirus P7 proteins known to be viral core proteins with nucleic acid binding activities. Interestingly, the N-terminal regions of PcV P3 (and corresponding P3 proteins of other chrysoviruses listed in Tables 2 and 3) share significant sequence similarity with comparable N-terminal regions of the putative RdRps encoded by chrysovirus dsRNA1s. The PcV P4 (and comparable proteins of other chrysoviruses) contains the motifs that form the conserved core of the ovarian tumor gene-like superfamily of predicted cysteine proteases.
Assignment of numbers 1–4 to PcV dsRNAs was made according to their decreasing size. Following the same criterion used for PcV, the dsRNAs associated with other chrysoviruses including Helminthosporium victoriae virus 145S (HvV145S), cherry chlorotic rusty spot associated chrysovirus (CCSRACV), Amasya cherry disease associated chrysovirus (ACDACV) and Cryphonectria nitschkei chrysovirus 1 (CnV1) (see below) were accordingly assigned the numbers 1–4. Sequence comparisons, however, indicated that dsRNAs 3 of Hv145SV, CCSRACV, ACDACV and CnV1 are in fact the counterparts of PcV dsRNA4 rather than dsRNA3. Likewise, dsRNA4 of these four chrysoviruses are the counterparts of PcV dsRNA3. Since PcV was the first chrysovirus to be characterized at the molecular level and to avoid confusion, the protein designations P3 and P4 as used for PcV are adopted and referred to as chryso-P3 and chryso-P4. Thus, whereas the chryso-P3 protein represents the gene product of PcV dsRNA3, it comprises the corresponding gene product of HvV145S dsRNA4, and so on.
The virion-associated RdRp catalyzes in vitro end-to-end conservative transcription of each dsRNA to produce mRNA by a conservative mechanism. Virions accumulate in the cytoplasm.
Biological properties
The viruses are associated with latent infections of their fungal hosts. There are no known natural vectors. The chrysoviruses are transmitted intracellularly during cell division and sporogenesis (vertical transmission) and by cell fusion following hyphal anastomosis between compatible fungal strains (horizontal transmission).
Antigenic properties
Virions are efficient immunogens.
Species demarcation criteria in the genus
The criteria to differentiate species in the genus Chrysovirus are:
- host range
- size of dsRNA segments
- length of 5′-UTR
- serological relationships.
List of species in the genus Chrysovirus
| Helminthosporium victoriae virus 145S |
|
|
| Helminthosporium victoriae virus 145S | [AF297176, AF297177, AF297178, AF297179] | (HvV145S) |
| Penicillium brevicompactum virus |
|
|
| Penicillum brevicompactum virus |
| (PbV) |
| Penicillium chrysogenum virus |
|
|
| Penicillium chrysogenum virus - ATTC 9480 | [AF296439, AF296440, AF296441, AF296442] | (PcV-ATCC9480) |
| Penicillium cyaneo-fulvum virus |
|
|
| Penicillium cyaneo-fulvum virus |
| (Pc-fV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Chrysovirus but have not been approved as species
| Amasya cherry disease associated chrysovirus | [AJ781163, AJ781164, AJ781165, AJ781166] | (ACDACV) |
| Cherry chlorotic rusty spot associated chrysovirus | [AJ781397, AJ781398, AJ781399, AJ781400] | (CCRSACV) |
| Cryphonectria nitschkei chrysovirus 1 | [DQ865186, DQ865187, DQ865188, DQ865189] | (CnV1) |
| Fusarium oxysporum chrysovirus 1 | [EF152346, EF152347, EF152348] | (FoCV1) |
List of unassigned species in the family Chrysoviridae
None reported.
Phylogenetic relationships within the family
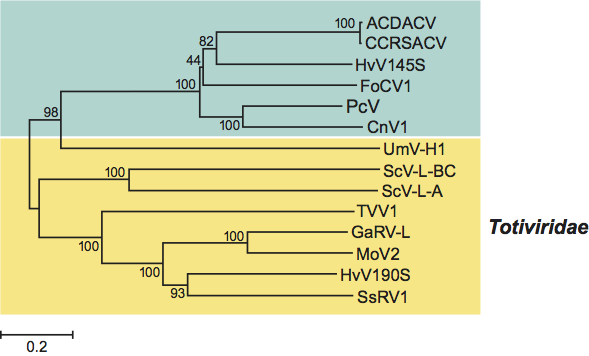
BLAST searches of PcV RdRp amino acid sequence showed that it has significantly high sequence similarity (37–50% identity and 55–67% sequence similarity) to the RdRps of CnV1, Hv145SV, FoCV1, ACDACV and CCRSACV. Phylogenetic analysis based on the complete deduced amino acid sequences of the RdRps of chrysoviruses and selected totiviruses confirmed that approved as well as probable members of the family Chrysoviridae (listed above) are most closely related to each other (Figure 3). These results suggest that the probable chrysoviruses CnV1, FoCV1, ACDACV and CCRSACV should be placed under the genus Chrysovirus in the family Chrysoviridae. High similarities (BLAST hits of 2e-11 or lower) were also found between PcV RdRp amino acid sequence and several members of the family Totiviridae including Ustilago maydis virus H1 (UmV-H1), Saccharomyces cerevisiae virus L-A (ScV-L-A), Trichomonas vaginalis virus (TVV) and Helminthosporium victoriae virus 190S (HvV190S). Interestingly, no significant hits were evident with any of the viruses in the family Partitiviridae, confirming the decision to remove the chrysoviruses from the family Partitiviridae and their placement in the newly created family Chrysoviridae.
Similarity with other taxa
Despite differences in the nature of the dsRNA genome between members in the families Chrysoviridae and Totiviridae (segmented versus nonsegmented genome), sequence and phylogenetic analyses of RdRp sequences suggest that chrysoviruses are most similar to viruses in the family Totiviridae (Figure 3).
Derivation of name
Chryso: from the specific epithet of Penicillium chrysogenum.
Further reading
Buck, K.W. and Grivan, R.F. (1977). Comparison of the biophysical and biochemical properties of Penicillium cyaneo-fulvum virus and Penicillium chrysogenum virus. J. Gen. Virol., 34, 145-154.
Castón, J.R., Ghabrial, S.A., Jiang, D., Rivas, G., Alfonso, C., Roca, R., Luque, D. and Carrascosa, J.L. (2003). Three-dimensional structure of Penicillium chrysogenum virus: a double-stranded RNA virus with a genuine T=1 capsid. J. Mol. Biol., 331, 417-431.
Covelli, L., Coutts, R.H.A., Di Serio, F., Citir, A., Acikgoz, S., Hernandez, C., Ragozzino. A. and Flores, R. (2004). Cherry chlorotic rusty spot and Amasya cherry diseases are associated with a complex pattern of mycoviral-like double-stranded RNAs. I. Characterization of a new species in the genus Chrysovirus. J. Gen. Virol., 85, 3389–3397.
Edmondson, S.P., Lang, D. and Gray, D.M. (1984). Evidence for sequence heterogeneity among double-stranded RNA segments of Penicillium chrysogenum mycovirus. J. Gen. Virol., 65, 1591-1599.
Ghabrial, S.A. (2008). Chrysoviruses. In: B.W.J. Mahy and M.H.V. Van Regenmortel (Eds.), Encyclopedia of Virology, 3rd edn, vol. 1. Elsevier, Oxford, pp. 503-513.
Ghabrial, S.A., Soldevila, A.I. and Havens, W.M. (2002). Molecular genetics of the viruses infecting the plant pathogenic fungus Helminthosporium victoriae. In: S. Tavantzis (Ed.), Molecular Biology of Double-stranded RNA: Concepts and Applications in Agriculture, Forestry and Medicine. CRC Press, Boca Raton, FL, pp. 213-236.
Jiang, D. and Ghabrial, S.A. (2004). Molecular characterization of Penicillium chrysogenum virus: reconsideration of the taxonomy of the genus Chrysovirus.J. Gen. Virol., 85, 2111-2121.
Liu, Y-C., Dynek, J. N., Hillman, B.I. and Milgroom, M.G. (2007). Diversity of viruses in Cryphonectria parasitica and C. nitschkei in Japan and China, and partial characterization of a new chrysovirus species. Mycol. Res., 111, 433-442.
Luque, D., González, J.M., Garriga, D., Ghabrial, S.A., Havens, W.M., Trus, B., Verdaguer, N., Carrascosa, J.L. and Castón, J.R. (2010). The T=1 capsid protein of Penicillium chrysogenum virus is formed by a repeated helix-rich core indicative of gene duplication. J. Virol. 84, 7256-7266.
Wood, H.A. and Bozarth, R.F. (1972). Properties of virus-like particles of Penicillium chrysogenum: one double-stranded RNA molecule per particle. Virology, 47, 604-609.
Contributed by
Ghabrial, S.A. and Castón, J.R.
Figures
Figure 1 (Left) Electron micrographs of Penicillium chrysogenum virus (PcV), the type species of the genus Chrysovirus. Samples were negatively stained in 2% uranyl acetate (lower panel) or prepared unstained and vitrified (upper panel); the bar represents 50 nm. (Right) Three-dimensional cryo-electron microscopy reconstruction of PcV virions at 8 resolution. Surface-shaded virion capsid viewed along an icosahedral twofold axis. Boundaries for two capsid proteins are outlined in black; each subunit has two similar domains (red and green), suggesting ancestral gene duplication. Icosahedral symmetry axes are indicated.
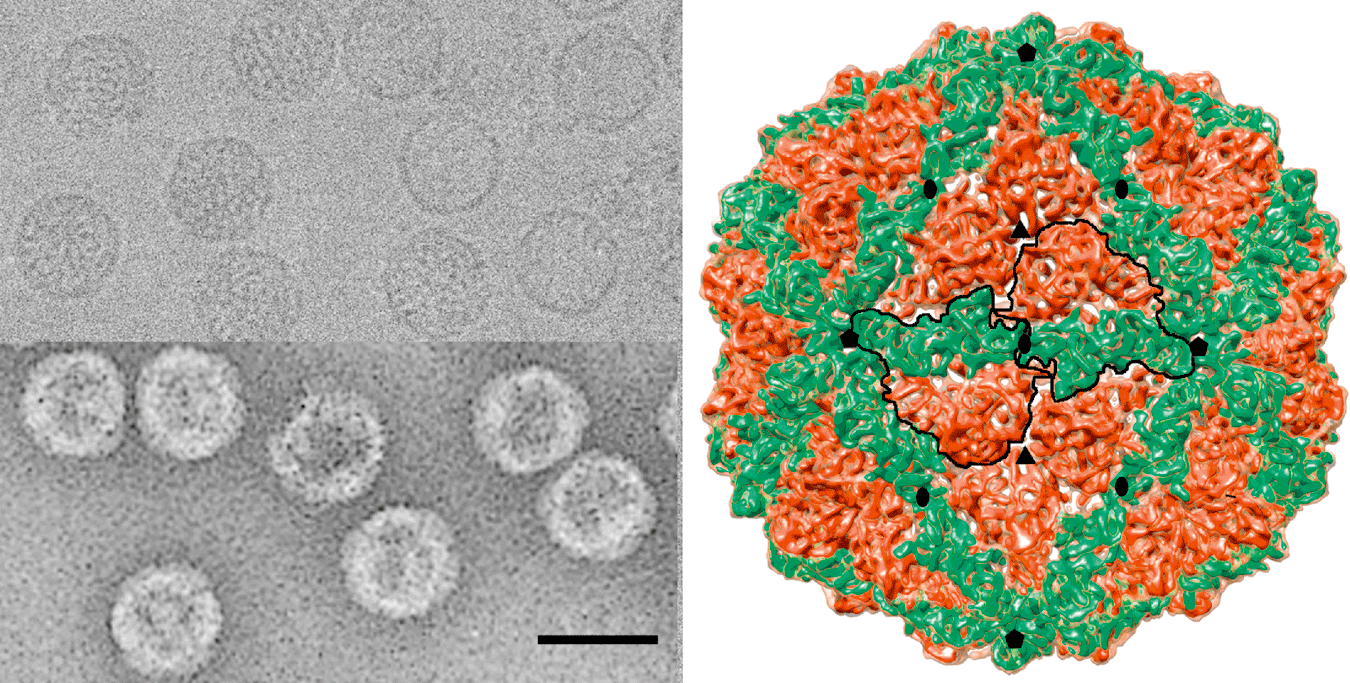
Figure 2 Genome organization of Penicillium chrysogenum virus (PcV). The genome consists of four dsRNA segments; each is monocistronic. The RdRp ORF (nt positions 145 to 3498 on dsRNA-1), the CP ORF (nt positions 158 to 3106 on dsRNA-2), the p3 ORF (nt positions 162 to 2900 on dsRNA-3) and the p4 ORF (nt positions 163 to 2706 on dsRNA-4) are represented by rectangular boxes.
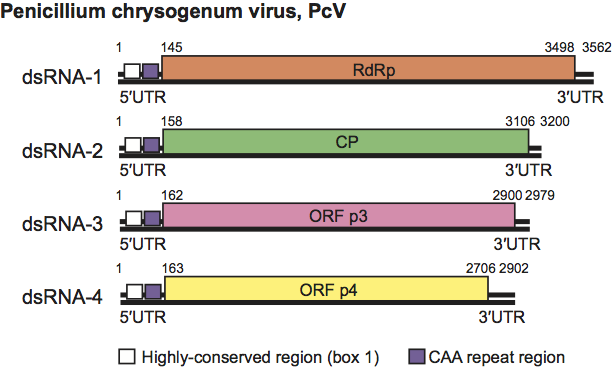
Figure 3 Neighbor-joining phylogenetic tree constructed based on the complete amino acid sequences of RdRps of members and probable members of the family Chrysoviridae and selected viruses from the family Totiviridae. The amino acid sequences were aligned using the program CLUSTAL W. See text for names and abbreviations of chrysoviruses. The following viruses in the family Totiviridae were included in the phylogenetic analysis (abbreviations in parenthesis): Gremmeniella abietina RNA virus L (GaRV-L), Helminthosporium victoriae virus 190S (HvV190S), Magnaporthe oryzea virus 2 (MoV2), Saccharomyces cerevisiae virus L-A (ScV-L-A), and ScV-L-BC, Sphaeropsis sapinea RNA virus 1 (SsRV1), Trichomonas vaginalis virus 1 (TVV1) and Ustilago maydis virus H1 (UmV-H1). The phylogenetic tree was generated using the MEGA4 phylogenetic package. Bootstrap values as a percent of 2000 replicates are indicated at each node.