Family: Birnaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Viruses of the family Birnaviridae are non-enveloped, single-shelled particles with a diameter of about 65 nm (Figure 1). The capsid follows a T=13 laevo icosahedral geometry and is made of a single capsid protein, VP2, clustered in trimers and forming 260 projections of about 4 nm at the surface of the particle.
Physicochemical and physical properties
Virion Mr is about 55×106, S20,W is 435S; buoyant density in CsCl is 1.33 g cm−3. Defective virions with interfering activity have been demonstrated to band at 1.30 g cm−3. Viruses are stable at pH 3–9, resistant to heat (60 °C, 1 h), ether and 1% SDS at 20 °C, pH 7.5 for 30 min.
Nucleic acid
Birnaviruses have a dsRNA genome which is made of two linear segments (A and B). The virions can package more than one complete genome copy that constitutes 8–10% of the particle by weight. The larger segment (A) is 3.1–3.6 kbp long, and the length of the smaller segment (B) ranges from 2.8 to 3.3 kbp, depending on the genus. The genome RNA sequences have a nucleotide composition of 53–58% G+C, with the exception of those of Rotifer birnavirus (RBV) and Drosophila X virus (DXV), which are 44–47% G+C. The segments are completely base-paired, and the plus-sense strand is covalently linked to a viral protein (VPg) at its 5′ terminus, but has neither a polyadenylation signal nor a terminal polyA tract at its 3′ end. Detailed characterization of the genome extremities have been carried out on Infectious bursal disease virus (IBDV) and Infectious pancreatic necrosis virus (IPNV). The first 30 nucleotides (nt) of the 5′-noncoding regions in the two segments display conserved motifs in both viruses. In contrast, sequences differ in the 3′-noncoding region of both segments. Inverted terminal repeats are present in the 5′- and 3′-noncoding regions of both segments. These motifs and repeats indicate the presence of cis-acting signals that are important for regulation of transcription, replication and selection of segments for packaging. Whereas the 5′-noncoding regions of both segments vary in length between 100 and 120 nt, the 3′-noncoding regions are about 60–150 nt in length. The 5′-noncoding regions are followed by one long ORF after the first AUG codon in segment B. Segment A also encodes a large ORF, which is generally preceded by a small, overlapping ORF. For Tellina virus 1 (TV-1), Blotched snakehead virus (BSNV) and DXV, the small ORF in segment A does not overlap the initiation codon of the large ORF. For RBV, there is no evidence for the presence of a small ORF in segment A.
Proteins
The polyprotein encoded by the large ORF in segment A is first processed during translation to generate preVP2, VP4 and VP3. Further processing of preVP2 occurs at its C-terminal domain to generate the mature capsid protein (VP2) and three or four peptides that remain in the particles. For IBDV, these peptides are 46, 7, 7 and 11 amino acid residues (aa) long. The capsid is formed by trimers of VP2 (417–442 aa). VP2 possesses a unique structural fold that is composed of two beta-barrels with a jelly-roll topology, oriented such that the beta-strands are tangential and radial to the virus particle. Inside the capsid, VP3 (238–309 aa) and the genomic RNA form thread-like ribonucleoprotein complexes that do not follow the icosahedral symmetry of the capsid. Different ratios of VP3 over VP2 have been reported in virions from members of different species.
The RNA-dependent RNA polymerase (RdRp), VP1 (844–1045 aa), is encoded by segment B. VP1 is found free in the viral particle and also covalently associated to the genome as VPg. The 2.5 Å resolution structure of IBDV VP1 reveals a characteristic rearrangement of motifs, from A–B–C to C–A–B, in the RNA polymerase catalytic palm domain, which is not found in viral RdRps from other dsRNA viruses. VP1 can guanylylate itself to produce VP1-pG and VP1-pGpG independently from its RNA polymerase activity. In the case of IBDV, VP1 has been shown to possess viral mRNA 5′-guanylyl transferase and capping activities.
VP4 (also called NS in IPNV, 212-244 aa) is a protease that cleaves its own N- and C- termini in the polyprotein and further processes preVP2. Its catalytic site is made of a serine-lysine dyad. The VP4 catalytic domain is structurally similar to the protease domain of bacterial ATP-dependent Lon proteases.
A nonstructural, positively charged polypeptide encoded by the small ORF of segment A has been designated VP5 (17 kDa in IBDV and 15 kDa in IPNV). This protein has been shown to be nonessential for replication of IBDV and IPNV. A second ORF, encoding an arginine-rich protein, has also been identified in genome segment A of DXV, BSNV and TV-1.
Lipids
None reported.
Carbohydrates
No N-linked glycosylation has been detected in any of the virion proteins. There is a report of O-linked glycosylation in IPNV VP2.
Genome organization and replication
As explained above, birnaviruses have a bipartite genome. The organization of segment A is illustrated in Figure 2. Generally, it contains two ORFs: ORF2 encoding a large polyprotein of about 105–120 kDa, and an overlapping (IPNV, IBDV and Drosophila birnavirus (DBV)) or internal (DXV, BSNV and TV-1) ORF1, which encodes a protein of 15–27 kDa. Positions of the polyprotein cleavage sites to generate preVP2, VP4 and VP3 during translation have been determined experimentally for IBDV, IPNV, BSNV, DXV and TV-1 (Figure 2). For BSNV and TV-1, an additional polypeptide, named X, is encoded between the preVP2 and the VP4 domains. The processing of preVP2 to generate VP2 and the structural peptides, which occurs during particle assembly, has been characterized for IBDV, IPNV, BSNV and TV-1. Sequence alignments allow the prediction of the preVP2 cleavage sites for other birnaviruses.
A single cycle of replication takes about 18–22 h for IPNV and 4–8 h for IBDV. The mode of entry of viruses into cells is not well understood, and the information is fragmentary. For IBDV binding at the cell surface, proteins such as heat shock protein 90 and α4β2 integrin have been proposed to serve as functional receptors in various types of chicken cells. Endosomal acidification is not a prerequisite for virus internalization in IPNV-infected cells. One of the small structural IBDV peptides, pep46 (a 46 aa amphiphilic peptide), and its homologs in other birnaviruses are able to induce pores in target membranes, suggesting a role in virus entry. After delivery into the cytoplasm, the virion RdRp becomes activated and produces two genome-length (24S) mRNA molecules from each of the 14S dsRNA genome segments. These mRNAs are capped, and they lack 3′ polyA sequences. Replicative intermediates have been identified in infected cells. Virus RNA is transcribed by a semi-conservative strand displacement mechanism in vitro. There is no information on minus strand RNA synthesis. The two mRNAs can be detected in infected cells by 3–4 h post infection (p.i.), and are synthesized in the same relative proportions throughout the replicative cycle (i.e. about twice as many A as B mRNA molecules). Virus-specific polypeptides can be detected at 4–5 h p.i. and are present in the same relative proportions until the end of the replication cycle. There are no specifically early or late proteins. The segment A mRNA is translated to yield a 105 kDa polyprotein that comprises the preVP2, VP4 (NS) and VP3 polypeptides, with the notable exception of BSNV and TV-1, which contain the X polypeptide between the preVP2 and VP4 domains (Figure 2). The VP4 protease co-translationally cleaves the polyprotein to generate three (or four in BSNV and TV-1) polypeptides (Figure 3). PreVP2 is later processed during virus assembly by a slow maturation cleavage to produce the mature VP2 and small structural peptides. This cleavage can be incomplete since traces of preVP2 are found in purified virus particles, although VP2 predominates. Virus assembly and maturation of the capsid protein preVP2/VP2 are concomitant and interdependent. In addition to VP4, the preVP2 processing requires the presence of VP3 and VP1. This requirement acts in favor of the existence of a large quaternary maturation complex formed by preVP2, VP4, VP3 and VP1. During infection, rigid tubes 55 nm in diameter are formed by preVP2. In the case of IBDV, additional tubules 25 nm in diameter, made of VP4, appear in late steps of the virus replication cycle.
The translation product of the 17 or 15 kDa ORF has been detected in IBDV or IPNV infected cells, respectively.
The mRNA from segment B is translated to a 94 kDa polypeptide that represents the viral RdRp (VP1, Figure 3). VP1 is found in virions in both a “free” and a genome-linked form (VPg). Virus particles assemble and accumulate in the cytoplasm. Encapsidation of the RdRp VP1 is mediated by its interaction with genome-associated protein VP3. The mechanism of virus release is unknown. In tissue culture, about half of the progeny virions remain cell-associated, and, depending on the multiplicity of infection, defective interfering particles are also formed.
Reverse genetics systems have been elaborated for IBDV and IPNV. In vitro transcribed viral cRNAs of segment A and B were found to be infectious, facilitating studies of birnavirus replication.
Antigenic properties
The capsid protein VP2 is the type-specific antigen and forms the virus-neutralizing epitopes. Anti-VP3 antibodies do not neutralize virus infectivity. There is no serological cross-reaction between the fish, avian and insect birnaviruses, or between the aquatic birnaviruses IPNV, BSNV, TV-1 and RBV.
Biological properties
The natural hosts of IPNV are salmonid fish, although this virus or viruses antigenically-related to it have also been isolated from other freshwater and marine fishes, as well as from bivalve molluscs (Tellina virus 2 (TV-2)). The virus is transmitted both vertically and horizontally, and there are no known vectors. The geographic distribution is worldwide. IPNV can cause epizootics resulting in high mortality in hatchery-reared salmonid fry and fingerlings. The virus causes necrotic lesions in the pancreas and is also found, without lesions, in other organs such as kidney, gonad, intestine and brain. It is believed that infected adult fish become lifelong carriers without exhibiting overt signs of infection.
The natural hosts of IBDV are chickens and turkeys. Rarely, IBDV has been isolated from ducks and other domestic fowl. The mode of transmission is horizontal, and there are no known vectors. IBDV has a worldwide distribution. The virus affects the bursa of Fabricius in young chicks, causing B lymphocyte deficiency. Death can occur between 3 and 10 weeks of age, and is associated with inflammation in the bursa of Fabricius, formation of immune complexes, depletion of complement, and clotting abnormalities.
Drosophila melanogaster populations are the natural host of DXV. The mode of transmission is horizontal and there are no known vectors. The geographic distribution is unknown. Infected fruitflies become sensitive to CO2. The target organs and histopathology are not known. DXV has also been isolated from populations of Culicoides spp.
BSNV was isolated from a cell line developed from a tropical fish species (Channa lucius), whereas TV-1 was identified in a bivalve mollusc (Tellina tenuis). RBV was isolated from a population collapse of the rotifer Brachionus plicatilis, which is cultivated for feeding the fry of marine fish in hatcheries. DBV was identified by deep sequencing of the small RNAs present in a cultured Drosophila melanogaster cell line.
Genus Aquabirnavirus
Type species Infectious pancreatic necrosis virus
Distinguishing features
Viruses in the genus thus far infect only fish, molluscs and crustaceans.
Biological properties
Aquabirnaviruses have been isolated from a variety of aquatic animals in freshwater, brackish or seawater environments. The ubiquitous nature of these agents and, in some cases, the lack of any association with disease has led to difficulty in assigning nomenclature. The first reports of isolation of IPNV were limited to epizootics in cultured brook trout (Salvalinus fontinalis). Soon IPNV was found to be responsible for disease in a variety of salmonid fish, including members of the genera Salmo, Salvalinus and Oncorhynchus. The virus has also been associated with disease in Japanese eels (Anguilla japonica) where it causes a nephritis, in menhaden (Brevoortia tryrannus) where it causes a “spinning disease,” and in yellowtail fingerlings (Seriola quinqueradiata) where it causes an ascites and cranial hemorrhage. In salmonid fish, IPNV causes acute gastroenteritis and destruction of the pancreas in the very young. A birnavirus has been associated with hematopoietic necrosis causing high mortalities in turbot (Scophthalmus maximus) with renal necrosis, and birnaviruses have been isolated from clams exhibiting darkened gills and gill necrosis. A non-typical apoptosis has been observed in cultured cells infected by IPNV.
Species demarcation criteria in the genus
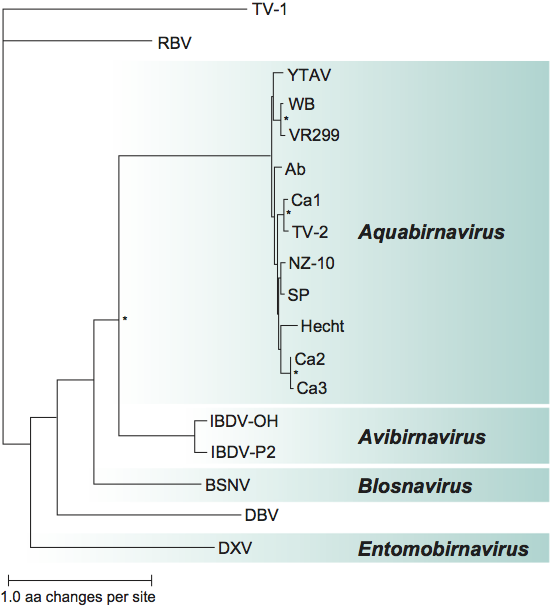
Three species of aquabirnaviruses are distinguished, primarily on the basis of host species. However, this classification was established before the current, broad species definition was adopted by the ICTV. The extremely close genetic relationship between the species, illustrated by Figure 4, suggests that there may be a case for combining them into one. Aquabirnaviruses do, nevertheless, display considerable antigenic diversity. Based on reciprocal neutralization assays using polyclonal antisera and immunoassays with monoclonal antibodies, the genus has been grouped into nine cross-reactive serotypes: A1 (type strain West Buxton), A2 (type strain Sp), A3 (type strain Ab), A4 (type strain Hecht), A5 (type Tellina virus-2), A6 (type strain Canada 1), A7 (type strain Canada 2), A8 (type strain Canada 3) and A9 (type strain VR299). Capsid protein sequences correlate well with serological classification and geographical distribution. Six genogroups were defined on the basis of sequence similarities: while genogroup 1 clusters serotypes A1 and A9, genogroup 2 corresponds to serotypes A7 and A8, genogroup 3 to serotype A3, genogroup 4 to serotypes A5 and A6, genogroup 5 to serotype A2 and genogroup 6 to serotype A4 (Figure 4). Further sequence characterization of aquabirnaviruses in Asia and Australasia evidenced the existence of an additional genogroup (type Yellowtail ascites virus) and an enlarged genogroup 5 including the strain NZ10 and relatives.
List of species in the genus Aquabirnavirus
| Infectious pancreatic necrosis virus |
|
|
| Infectious pancreatic necrosis virus - West Buxton | [A:AF078668; B:AF078669] | (IPNV-WB) |
| Tellina virus |
|
|
| Tellina virus 2 | [A:AF342730] | (TV-2) |
| Yellowtail ascites virus |
|
|
| Yellowtail ascites virus - Y-6 | [A:AB006783; B:AY129662] | (YTAV-Y-6) |
Species names are in italic script; names of strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Aquabirnavirus but have not been approved as species
| Marine birnavirus - AY-98 | [B: AY123970] | (MABV-AY-98) |
| Marine birnavirus - H-1 | [B: AY129665] | (MABV-H-1) |
Genus Avibirnavirus
Type species Infectious bursal disease virus
Distinguishing features
Viruses in the genus thus far infect only birds.
Biological properties
IBDV causes an immunosuppressive disease in chickens by destruction of B lymphocytes in the bursa of Fabricius. Apoptosis has also been observed in this and other lymphoid organs. VP5 inhibits apoptosis at the early stage of viral infection in chicken embryonic fibroblast cells, whereas VP2 induces apoptosis in transfected mammalian cells. The latter finding correlates with evidence of apoptosis and B cell death in chickens infected with IBDV. The rapid depletion of B cells in the bursa of Fabricius leads to immunosuppression and increased susceptibility to other infections and diseases. The virus is highly contagious and is of major importance to the poultry industry worldwide. Two serotypes (1 and 2) of IBDV have been identified by cross-neutralization assays. Serotype 1 strains are pathogenic in chickens, whereas serotype 2 strains are nonpathogenic.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Avibirnavirus
| Infectious bursal disease virus |
|
|
| Infectious bursal disease virus - strain P2 | [A: X84034; B:X84035] | (IBDV-P2) |
Species names are in italic script; names of strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Avibirnavirus but have not been approved as species
None reported.
Genus Blosnavirus
Type species Blotched snakehead virus
Distinguishing features
Viruses in the genus thus far infect only fish.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Blosnavirus
| Blotched snakehead virus |
|
|
| Blotched snakehead virus | [A:AJ459382, B:AJ459383] | (BSNV) |
Species names are in italic script; names of strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Blosnavirus but have not been approved as species
None reported
Genus Entomobirnavirus
Type species Drosophila X virus
Distinguishing features
Viruses in this genus thus far infect only insects.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Entomobirnavirus
| Drosophila X virus |
|
|
| Drosophila X virus | [A:U60650, B:AF196645] | (DXV) |
Species names are in italic script; names of strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Entomobirnavirus but have not been approved as species
None reported.
List of other related viruses which may be members of the family Birnaviridae but have not been approved as species
| Rotifer birnavirus | [A:FM995220, B:FM995221] | (RBV) |
| Tellina virus 1 | [A:AJ920335, B :AJ920336] | (TV-1) |
| Drosophila birnavirus | [A:GQ342962, B:GQ342963] | (DBV) |
Phylogenetic relationships within the family Birnaviridae
See Figure 4.
Similarity with other taxa
Birnaviruses share no nucleic acid sequence similarity with other taxa. In the same way, the capsid protein VP2 has no sequence similarities with corresponding capsid proteins of any other virus family. However, the crystal structures of IBDV and IPNV VP2 shows that VP2 is folded into three distinct domains (base, shell and projection) disposed radially in the virus particle, the base and shell domains displaying high structural similarities with the capsid proteins of viruses belonging to the families Nodaviridae and Tetraviridae, which contain positive-strand ssRNA viruses. The VP4 protease has sequence and structural homologies with the Ser-Lys catalytic protease domain of bacterial ATP-dependent Lon proteases. Interestingly, the A-B-C to C-A-B motif rearrangement in the VP1 RdRps of birnaviruses, noted above, is also shared by the RdRps of Thosea asigna virus and Euprosterna elaeasa virus, insect viruses that have positive strain ssRNA genomes (genus Betatetravirus, family Tetraviridae), but not by RdRps of other dsRNA viruses. These unusual RdRps form a minor and deeply separated cluster in the viral RdRp phylogenetic tree. The structural relationships among the capsid proteins and the RdRps of birnavirus and viruses belonging to the family Tetraviridae suggest evolutionary links between positive-strand ssRNA and dsRNA viruses.
Derivation of names
Aqua: from Latin aqua, “water”.
Avi: from Latin avis, “bird”.
Birna: from Latin prefix bi, “two”, signifying the bisegmented nature of the viral genome as well as the presence of dsRNA; and rna from ribo nucleic acid, indicating the nature of the viral genome.
Entomo: from Greek entomon, “insect”.
Blosna: from blotched snakehead virus.
Further reading
Birghan, C., Mundt, E. and Gorbalenya, A.E. (2000). A non-canonical lon proteinase lacking the ATPase domain employs the ser-Lys catalytic dyad to exercise broad control over the life cycle of a double-stranded RNA virus. EMBO J., 19, 114-123.
Blake, S., Ma, J.-Y., Caporale, D. A., Jairath, S. and Nicholson, B. L. (2001). Phylogenetic relationships of aquatic birnaviruses based on deduced amino acid sequences of genome segment A cDNA. Dis. Aquat. Organ., 45, 89-102.
Coulibaly, F., Chevalier, C., Gutsche, I., Pous, J., Navaza, J., Bressanelli, S., Delmas, B. and Rey, F. (2005). The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell, 120, 761-772.
Gorbalenya, A.E., Pringle, F.M., Zeddam, J.L., Luke, B.T., Cameron, C.E., Kalmakoff, J., Hanzlik, T.N., Gordon, K.H. and Ward, V.K. (2002). The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol., 324, 47-62.
Lombardo, E., Maraver, A., Caston, J.R., Rivera, J., Fernandez-Arias, A., Serrano, A., Carrascosa, J.L. and Rodriguez, J.F. (1999). VP1, the putative RNA-dependent RNA polymerase of infectious bursal disease virus, forms complexes with the capsid protein VP3, leading to efficient encapsidation into virus-like particles. J. Virol., 73, 6973-6983.
Luque, D., Rivas, G., Alfonso, C., Carrascosa, J.L., Rodriguez, J.F., and Caston, J.R. (2009). Infectious bursal disease virus is an icosahedral polyploid dsRNA virus. Proc. Natl Acad. Sci., USA, 106, 2148-2152.
Mundt, E. and Vakharia, V.N. (1996). Synthetic transcripts of double-stranded birnavirus genome are infectious. Proc. Natl Acad. Sci., USA, 93, 11131-11136.
Nobiron, I., Galloux, M., Henry, C., Torhy, C., Boudinot, P., Lejal, N., Da Costa, B. and Delmas, B. (2008). Genome and polypeptides characterization of Tellina virus 1 reveals a fifth genetic cluster in the Birnaviridae family. Virology, 371, 350-361.
Pan, J., Vakharia, V.N. and Tao, Y.J. (2007). The structure of a birnavirus polymerase reveals a distinct active site topology. Proc. Natl Acad. Sci., USA, 104, 7385-7390.
Wu, Q., Luo, Y., Lu, R., Lau, N., Lai, E.C., Li, W.-X. and Ding, S.-W. (2010). Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl Acad. Sci., USA, 107, 1606-1611.
Contributed by
Delmas, B., Mundt, E., Vakharia, V.N. and Wu, J.L.
Figures
Figure 1 (Top) Negative contrast electron micrograph of infectious bursal disease virus (IBDV) particles (courtesy of J. Lepault). Bar represents 100 nm. (Bottom left) A three-dimensional model of the IBDV virion derived from X-ray crystallography (courtesy of F. Rey). (Bottom right) Diagrammatic representation of an IBDV particle, which has a single T=13 icosahedral shell.
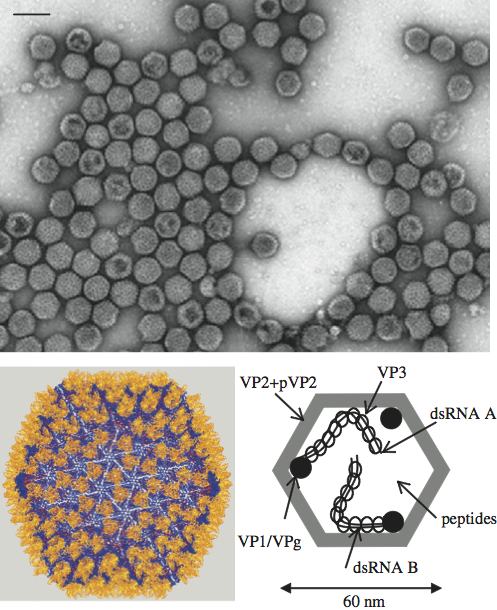
Figure 2 Schematic representation of the gene arrangement in genome segment A of birnaviruses representative of the family: infectious bursal disease virus (IBDV), blotched snakehead virus (BSNV), infectious pancreatic necrosis virus (IPNV), Drosophila birnavirus (DBV), Tellina virus 1 (TV-1), rotifer birnavirus (RBV) and Drosophila X virus (DXV). Polyprotein (ORF2) cleavage sites are indicated by a vertical bar and their positions by the P1/P1 number. The location and size of the small ORFs (ORF1) are indicated below the polyprotein ORF. Numbers in parentheses at the 5 ends indicate the length of the 5-non-coding region.
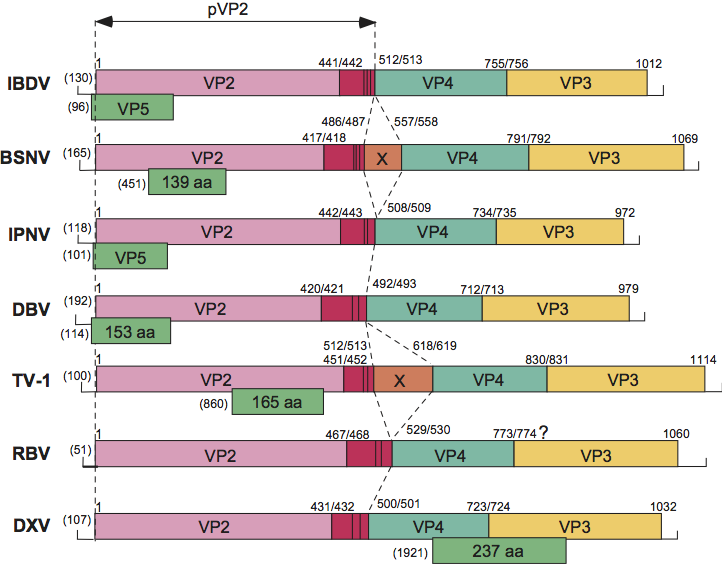
Figure 3 Schematic representation of the genome of infectious bursal disease virus (IBDV) illustrating processing of the encoded proteins. Numbers in parentheses indicate the nucleotide lengths of the two genomic segments.
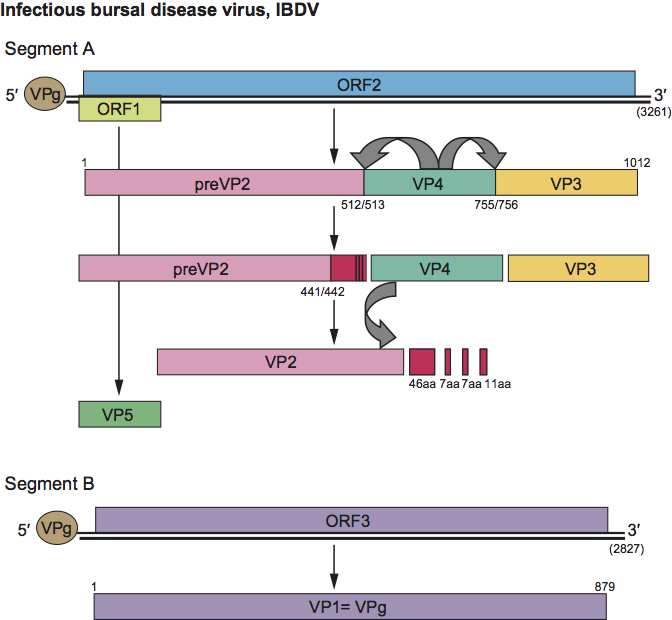
Figure 4 A distance tree representing the phylogenetic relationships of VP2 in the various genera and genetic clusters of the family. Alignment was performed with MUSCLE and phylogenetic analysis with PhyML using default parameters. Internal branches with bootstrap support above 80% are highlighted. The tree was inferred by maximum likelihood on the amino-acid sequences of the capsid protein. Tellina virus 1 (TV-1), Drosophila birnavirus (DBV) and rotifer birnavirus (RBV) are separate from each other and from the four genera. The protein accession numbers used for comparison were (top to bottom): TV-1: CAI74981, RBV: CAX33877, YTAV: BAA25005, WB: AF342727, VR299: AF343572, Ab: AF342729, Ca1: AF342732, TV-2: AF342731, NZ-10: ACG56371, SP: AF342728, Hecht: AF342730, Ca2: AF342733, Ca3: AF342734, IBDV-OH: AAC55351, IBDV-P2: CAA58851, BSNV: AJ459382, DBV: ACU32790, and DXV: AAB16798.