Family: Tectiviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Tectivirus
Type species Enterobacteria phage PRD1
Distinguishing features
Characteristic features of the tectiviruses are a linear dsDNA genome and a protein-rich internal membrane, which is enclosed in an icosahedral protein capsid. The genome has around 100 bp long inverted terminal repeats (ITR) and covalently linked 5′-terminal proteins used in replication as primers. Upon infection the viral membrane is transformed to a tubular structure used in genome delivery.
Virion properties
Morphology
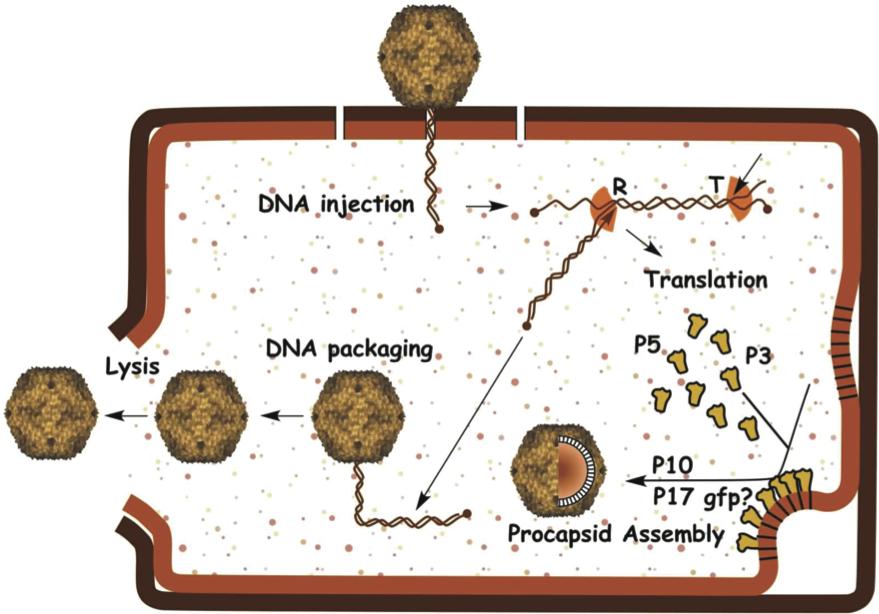
Virions are icosahedra, have no external envelope, and measure 66 nm from facet to facet (Figure 1). Capsids have flexible spikes extending about 20 nm from the virion vertices. The capsid of Enterobacteria phage PRD1 (PRD1) is constructed of 240 major capsid protein (P3) trimers that form a pseudo T=25 lattice. Protein P3 contains two beta-barrels and forms very tight trimers. Underneath the capsomers the minor coat protein P30 dimers stretch out from one vertex to another cementing the P3 facets together. The spikes are formed by two proteins (P2 and P5) extending from the penton protein (P31) and are used for receptor recognition. One virion vertex is different and used for the phage DNA packaging (Figure 1). The packaging vertex contains packaging ATPase P9 and three other proteins (P6, P20 and P22). The capsid encloses an inner membrane formed of approximately equal amounts of virus-encoded proteins and lipids derived from host cell plasma membrane. The DNA is coiled within this membrane. Virions are normally tailless, but produce tail-like tubes of about 60×10 nm upon DNA release or after chloroform treatment (Figure 1).
Physicochemical and physical properties
The virion Mr is about 66×106, the S20,w is 357–416S, and the buoyant density in CsCl is about 1.29 g cm−3. Virions are usually stable at pH 5–8. Infectivity is sensitive to organic solvents and detergents.
Nucleic acid
Virion contains a single molecule of linear dsDNA of about 15 kb. The DNA corresponds to 14–15% of particle weight and the G+C content is 48%. The complete DNA sequences of phage PRD1 and five closely related phages (Enterobacteria phages PR3, PR4, PR5, PR772 and L17) as well as Bacillus phages Bam35, GIL16, GIL01 and AP50 have been determined. The 100 bp long ITRs of PRD1 are 100% identical.
Proteins
PRD1 proteins and their functions are listed in Table 1. The proteins of other enterobacterial tectiviruses are closely related to PRD1. Tectiviruses infecting members of the genus Bacillus (Bam35, GIL16, GIL01, and AP50) contain approximately the same number of proteins as PRD1.
Table 1 Enterobacteria phage PRD1 (PRD1) proteins and their functions
| Phage protein | Gene | Mutant | Mass (kDa) | Function | Location |
| P1 | I | + | 63.3 | DNA polymerase | Cytoplasm |
| P2 | II | + | 63.7 | Receptor binding | Vertices |
| P3 | III | + | 43.1 | Major capsid protein | Capsid |
| P5 | V | + | 34.2 | Spike | Vertices |
| P6 | VI | + | 17.6 | Packaging | Unique vertex |
| P7 | VII | + | 27.1 | DNA entry, transglycosylase | Viral membrane |
| P8 | VIII | + | 29.6 | Genome terminal protein | Viral DNA |
| P9 | IX | + | 25.8 | DNA packaging | Unique vertex |
| P10 | X | + | 20.6 | Assembly | Host plasma membrane |
| P11 | XI | + | 22.2 | DNA entry | Membrane surface |
| P12 | XII | + | 16.6 | ssDNA binding | Cytoplasm |
| P14 | XIV | + | 15.0 | DNA entry | Vertices |
| P15 | XV | + | 17.3 | Lytic muramidase | Cytoplasm? |
| P16 | XVI | + | 12.6 | Infectivity | Viral membrane |
| P17 | XVII | + | 9.5 | Assembly | Cytoplasm |
| P18 | XVIII | + | 9.8 | DNA entry | Viral membrane |
| P19 | XIX | + | 10.5 | ssDNA binding | Cytoplasm |
| P20 | XX | + | 4.7 | DNA packaging | Unique vertex |
| P22 | XXII | + | 5.5 | DNA packaging | Unique vertex |
| P30 | XXX | + | 9.1 | Capsid stability | Capsid |
| P31 | XXXI | + | 13.7 | Penton | Capsid |
| P32 | XXXII | + | 5.4 | DNA entry | Viral membrane |
| P33 | XXXIII | − | 7.5 | Assembly | Cytoplasm |
| P34 | XXXIV | − | 6.7 | ? | Viral membrane |
| P35 | XXXV | + | 12.8 | Holin | Host plasma membrane |
| P36 | XXXVI | + | 12.6 | Lysis | Host plasma membrane |
| P37 | XXXVII | + | 10.1 | Lysis | Host plasma membrane |
Lipids
Virions contain about 15% lipids by weight. Lipids form an internal membrane with virus-specific proteins. Lipids constitute about 60% of the inner membrane. In PRD1, lipids form a bilayer and seem to be in a liquid crystalline phase. Phospholipid content (52% phosphatidylethanolamine and 43% phosphatidylglycerol, 5% cardiolipin) deviates from the host plasma membrane such that the phosphatidylglycerol is enriched. The fatty acid composition is close to that of the host.
Carbohydrates
Not detected.
Genome organization and replication
The PRD1 genome is a linear dsDNA molecule of 14,927 bp with proteins covalently linked to both 5’-termini. Replication is protein-primed, proceeds by strand displacement and can start at both ends of the genome as the ITRs contain sites for the replication initiation. A total of 27 identified genes are distributed in both strands and organized into five operons. Both ends of the genome contain early genes involved in replication and regulation of gene expression (Figure 2). The genes located in the three late operons encode structural proteins and proteins involved in assembly and lysis.
After DNA entry, replication and transcription, capsid proteins polymerize in the cytoplasm, whereas membrane-associated proteins are inserted into the host plasma membrane (Figure 3). With the help of nonstructural virion-encoded assembly factors and the coat-forming proteins, a virus-specific lipoprotein membrane obtains an outer protein shell leading to the formation of a procapsid. This is translocated to the interior of the cell. The linear genome is packaged into the procapsid through a unique packaging vertex. Mature virions are released by lysis of the host cell (Figure 3).
Antigenic properties
No information available.
Biological properties
Enterobacterial phages are virulent. The tail-like membranous tube probably acts as a DNA injection device. Phages Bam35 and AP50 and a number of additional isolates are specific for Bacillusspecies and are temperate. Thermus phage P37-14 and a number of similar isolates are specific for Thermus and are found in volcanic hot springs.
Species demarcation criteria in the genus
The sequence comparison of tectiviruses infecting Enterobacteria (phage PRD1) and Bacillus (phage Bam35) reveals very little identity, but genome organization and location of key genes are similar. The Thermus phage hosts are thermophilic and the other tectivirus hosts are mesophilic.
List of species in the genus Tectivirus
| Bacillus phage AP50 |
|
|
| Bacillus phage AP50 | [EU408779] | (AP50) |
| Bacillus phage Bam35 |
|
|
| Bacillus phage Bam35 | [AY257527] | (Bam35) |
| Enterobacteria phage PRD1 |
|
|
| Enterobacteria phage PRD1 | [AY848689] | (PRD1) |
| Thermus phage P37-14 |
|
|
| Thermus phage P37-14 |
| (P37-14) |
Species names are in italic script; names of strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Tectivirus but have not been approved as species
| Enterobacteria phage L17 | [AY848684] | (L17) |
| Enterobacteria phage PR3 | [AY848685] | (PR3) |
| Enterobacteria phage PR4 | [AY848686] | (PR4) |
| Enterobacteria phage PR5 | [AY848687] | (PR5) |
| Enterobacteria phage PR772 | [AY848688] | (PR772) |
| Bacillus phage GIL16 | [AY701338] | (GIL16) |
| Bacillus phage GIL01 | [AJ536073] | (GIL01) |
List of unassigned species in the family Tectiviridae
| Bacillus phage phiNS11 |
|
|
| Bacillus phage phiNS11 |
| (phiNS11) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
Phylogenetic relationships within the family
The Enterobacteria phages (PRD1-types) show nucleotide sequence similarity between 91.9% and 99.8%. The Bacillus phages (Bam35-types) are more diverse in sequence (59–99% identity). Bam35- and PRD1-type phages most probably share a common ancestor.
Similarity with other taxa
PRD1, and presumably other tectiviruses, have many features in common with adenoviruses. These include: (1) a linear dsDNA genome, (2) genomic inverted terminal repeats, (3) protein-primed initiation of replication, (4) type B (E. coli Pol II) DNA polymerase, (5) trimeric capsid proteins that contains two beta-barrels and form hexagonal structures, (6) arrangement of the capsid protein in a pseudo T=25 lattice, (7) five-fold protein (penton) fold and (8) receptor-binding spikes at capsid vertices. The same principal capsid architecture and capsid protein fold have also been described in eukaryotic Paramecium bursaria Chlorella virus 1 (family Phycodnaviridae), Chilo iridescent virus (family Iridoviridae), archaeal Sulfolobus turreted icosahedral virus (unassigned family) and bacterial Pseudoalteromonas phage PM2 (family Corticoviridae).
Derivation of name
Tecti: from Latin tectus, “covered”.
Further reading
Abrescia, N.G., Cockburn, J.J., Grimes, J.M., Sutton, G.C., Diprose, J.M., Butcher, S.J., Fuller, S.D., San Martin, C., Burnett, R.M., Stuart, D.I, Bamford, D.H. and Bamford, J.K. (2004). Insights into assembly from structural analysis of bacteriophage PRD1. Nature, 432, 68-74.
Benson, S.D., Bamford, J.K.H., Bamford, D.H. and Burnett, R.M. (1999). Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell, 98, 825-833.
Cockburn, J.J., Abrescia, N.G., Grimes, J.M., Sutton, G.C., Diprose, J.M., Benevides, J.M., Thomas, G.J. Jr., Bamford, J.K.H., Bamford, D.H. and Stuart, D.I. (2004). Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature, 432, 122-125.
Grahn, A.M., Butcher, S.J., Bamford, J.K.H., and Bamford, D.H. (2006). PRD1: Dissecting the genome, structure, and entry. In: R. Calendar (Eds.), The Bacteriophages. Oxford University Press, New York, pp. 161-170.
Laurinmäki, P.A., Huiskonen, J.T., Bamford, D.H. and Butcher, S.J. (2005). Membrane proteins modulate the bilayer curvature in the bacterial virus Bam35. Structure, 13, 1819-1828.
Ravantti, J.J., Gaidelyte, A., Bamford, D.H. and Bamford, J.K.H. (2003). Comparative analysis of bacterial viruses Bam35, infecting a gram-positive host, and PRD1 infecting gram-negative hosts, demonstrate a viral lineage. Virology, 313, 401-414.
Saren, A.M., Ravantti, J.J., Benson, S.D., Burnett, R.M., Paulin, L., Bamford, D.H. and Bamford, J.K.H. (2005). A snapshot of viral evolution from genome analysis of the Tectiviridae family. J. Mol. Biol., 350, 427-440.
Contributed by
Oksanen, H.M and Bamford, D.H.
Figures
Figure 1 (Left) X-ray crystallography-based structure of the virion of Enterobacteria phage PRD1 (pseudo T=25) viewed down two-fold axis of symmetry. (Middle) Schematic picture of the virion. (Right) Negative stain electron micrograph of phage PRD1 particles. The inset shows a particle with a protruding membranous tail-like structure. The bar represents 100 nm.
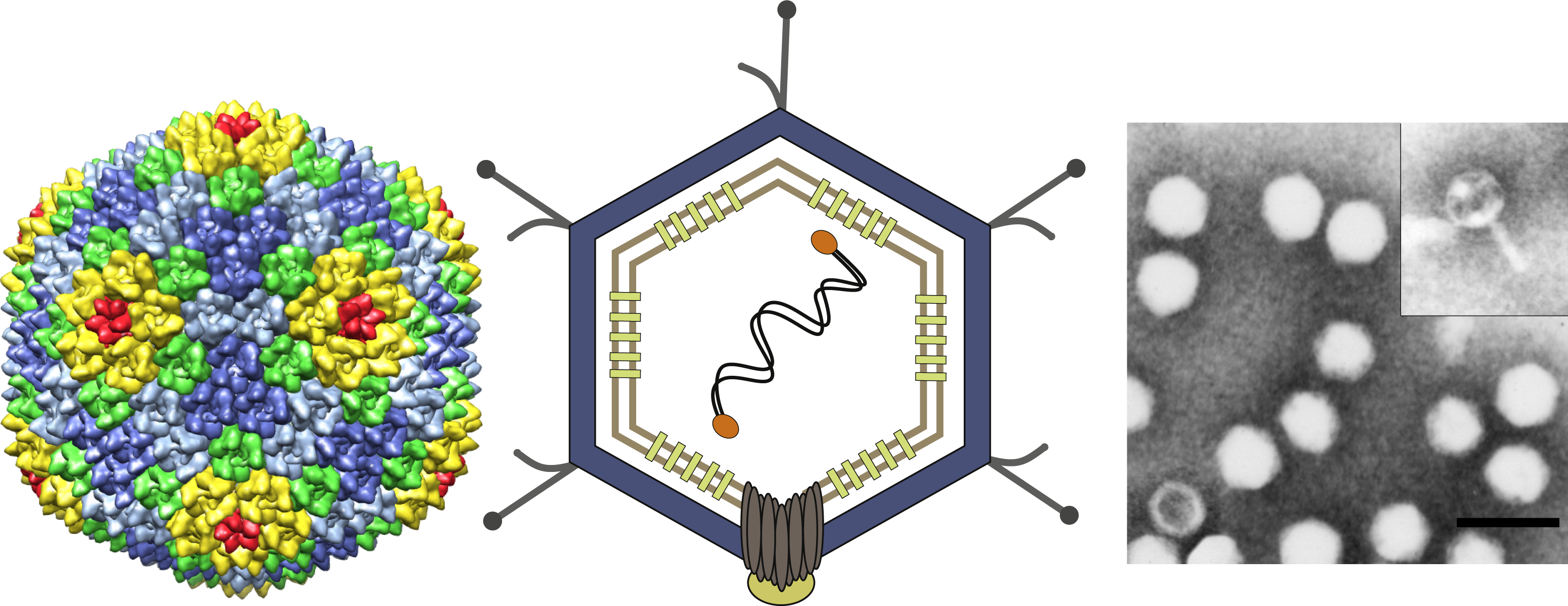
Figure 2 The genome of Enterobacteria phage PRD1 (PRD1) indicating the organization of genes and promoters (P) and their reading direction. Gene and protein nomenclature are correlated so that each protein has the same number (Arabic numeral) as the gene (Roman numeral). ORFs with no confirmed protein product are indicated by lower case letters. (Top) Order of genes; genes XIX and XII are on one strand and all other genes are on the other strand. The different colors indicate the ORFs (yellow) and the genes coding structural proteins (blue), nonstructural proteins (orange) and proteins involved in lysis (green). (Bottom) Promoters (P), terminators (lollipops) and operons (O) and their reading directions; E=early, L=late.
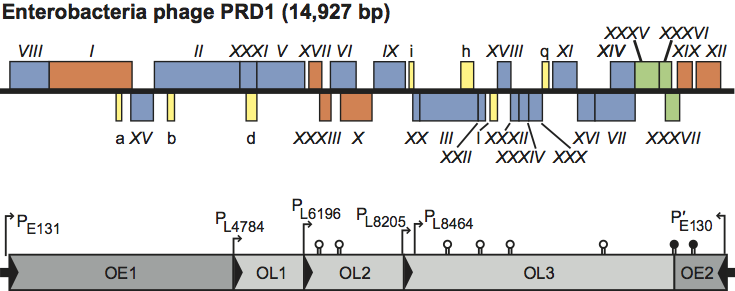
Figure 3 Diagram of the life cycle of Enterobacteria phage PRD1; for details see the text.