Family Siphoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Distinguishing features
Virions have long, non-contractile, thin tails (65–570×7–10 nm) which are often flexible. Tails are built of stacked disks of six subunits. Heads and tails are assembled separately. Genera are differentiated by genome organization, mechanisms of DNA packaging and presence or absence of DNA polymerases.
Genus “Lambda-like viruses”
Type species Enterobacteria phage lambda
Distinguishing features
The DNA has cohesive ends and is packaged as a unit-size filament.
Virion properties
Morphology
Phage heads are icosahedra, about 60 nm in diameter, and consist of 72 capsomers (60 hexamers, 12 pentamers, T=7). Tails are flexible, 150×8 nm, and have a short terminal fiber and four long, jointed fibers attached subterminally (Figure 1). The latter fibers are absent in most laboratory strains of the phage.
Physicochemical and physical properties
Virion Mr is about 60 ×106, buoyant density in CsCl is 1.50 g cm−3, and the S20,w is about 390S. Infectivity is chloroform- and ether-resistant.
Nucleic acid
The phage λ genome is 48,503 bp in size, corresponding to 54% of particle weight, has 52% G+C. The DNA has cohesive ends and is non-permuted.
Proteins
Virions contain about 14 structural proteins (11–130 kDa), including 415 copies each of major capsid proteins E and D (38 and 11 kDa, respectively).
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genome includes about 70 genes and has cohesive ends (Figure 2). Related functions cluster together. The infecting DNA circularizes and either replicates or integrates into the host genome. Transcription starts in the immunity region and proceeds in three waves. Bidirectional DNA replication as a Θ (theta) structure, starting from a single site, is followed by unidirectional replication via a rolling-circle mechanism. There is no breakdown of host DNA. Proheads are frequent in lysates.
Biological properties
Phages are temperate and apparently specific for enterobacteria. Phages generally integrate at specific sites and are UV-inducible.
Species demarcation criteria in the genus
Species are distinguished by different combinations of alleles of genes encoding head proteins, homologous recombination proteins, and DNA replication proteins, in the context of very similar genome organization.
List of species in the genus “Lambda-like viruses”
| Enterobacteria phage HK022 |
|
|
| Enterobacteria phage HK022 | [AF069308] | (HK022) |
| Enterobacteria phage HK97 |
|
|
| Enterobacteria phage HK97 | [AF069529] | (HK97) |
| Enterobacteria phage lambda |
|
|
| Enterobacteria phage λ | [J02459] | (λ) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “Lambda-like viruses” but have not been approved as species
| Enterobacteria phage PA-2 | (PA-2) |
| Enterobacteria phage FD328 | (FD328) |
| Enterobacteria phage 80 | (80) |
| Rhizobium phage 16-6-2 | (16-6-2) |
Genus “T1-like viruses”
Type species Enterobacteria phage T1
Distinguishing features
Tails are extremely flexible. Phage DNA has pac sites and is terminally redundant and circularly permuted.
Virion properties
Morphology
Virions have icosahedral heads of about 60 nm and extremely flexible tails of 151×8 nm, with four short, kinked, terminal fibers. The flexible nature of the tail is best seen after phosphotungstate staining.
Physicochemical and physical properties
Virion buoyant density in CsC1 is 1.5 g cm−3. Virion infectivity is stable to drying.
Nucleic acid
Genomes are about 49 kbp and have a G+C content about 48%.
Proteins
Virions contain at least 14 proteins, including two major head proteins (26 and 33 kDa).
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genetic map is linear and comprises 36–41 genes; related functions cluster together. The genome is circularly permuted and terminally redundant (2.8 kbp or 6% of genome), and includes a recombinational hot spot. Host syntheses are inhibited after infection. Little is known about the mechanism of Enterobacteria phage T1 replication. Progeny DNA is cut from concatemers at pac sites and packaged by a headfull mechanism.
Biological properties
Phages are virulent, can carry out generalized transduction and infect enterobacteria.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus “T1-like viruses”
| Enterobacteria phage T1 |
|
|
| Enterobacteria phage T1 | [AY216660] | (T1) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “T1-like viruses” but have not been approved as species
| Enterobacteria phage 102 | (102) |
| Enterobacteria phage 103 | (103) |
| Enterobacteria phage 150 | (150) |
| Enterobacteria phage 168 | (168) |
| Enterobacteria phage 174 | (174) |
| Enterobacteria phage b4 | (b4) |
| Enterobacteria phage D20 | (D20) |
| Enterobacteria phage fg | (fg) |
| Enterobacteria phage Hi | (Hi) |
| Enterobacteria phage UC-1 | (UC-1) |
Genus “T5-like viruses”
Type species Enterobacteria phage T5
Distinguishing features
Virions have large heads and long, kinked tail fibers. The DNA has five single-stranded gaps, large terminal repetitions, codes for a type A (E. coli Pol I) DNA polymerase, and is injected in two steps.
Virion properties
Morphology
Virions have icosahedral heads about 80 nm in diameter. Tails measure 180×9 nm in Enterobacteria phage T5 (T5) and 160×9 nm in Vibrio phage 149 (type IV), have a subterminal disk with three kinked fibers about 120 nm in length, and a conical tip with a single, straight, 50 nm long fiber. Vibrio phage 149 tail fibers have terminal knobs.
Physicochemical and physical properties
Virion Mr is 114×106, buoyant density in CsC1 is 1.53 g cm−3, and S20,w is 608S.
Nucleic acid
Genomes are about 121 kbp, corresponding to 62% of particle weight, and have a G+C content of 44%.
Proteins
Virions contain at least 15 structural proteins (15.5–125 kDa), including about 775 copies of the major capsid protein (44 kDa). Type A (E. coli Pol I) DNA polymerase is encoded.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genetic map is linear. The genome includes at least 80 genes, is divided into five regions, is non-permuted, and has a large terminal repetition of about 10 kbp (8.5% of genome) and five single stranded gaps at specific sites. It has neither cos nor pac sites. Pre-early, early and late genes cluster together. Only 8% of DNA is injected immediately after adsorption; the rest follows after 3–4 minutes. Transcription involves modification of host RNA polymerase by phage gene products. DNA replication follows a bidirectional or a rolling-circle mechanism or both. Concatemers are produced.
Biological properties
Infection is virulent. Known phages of the genus infect enterobacteria and vibrios.
Species demarcation criteria in the genus
Species differ by host range.
List of species in the genus “T5-like viruses”
| Enterobacteria phage T5 |
|
|
| Enterobacteria phage T5 | [AY543070] | (T5) |
| Vibrio phage 149 (type IV) |
|
|
| Vibrio phage 149 (type IV) |
| (149) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “T5-like viruses” but have not been approved as species
| Enterobacteria phage BF23 | (BF23) |
| Enterobacteria phage PB | (PB) |
| Enterobacteria phage San 2 | (San2) |
Genus “L5-like viruses”
Type species Mycobacterium phage L5
Distinguishing features
Phage DNA has cos sites and codes for a type A (E. coli Pol I) DNA polymerase.
Virion properties
Morphology
Virions have isometric heads about 60 nm in diameter and flexible tails of 135×8 nm with a terminal knob and a single short fiber.
Physicochemical and physical properties
Virion Mr is 116×106, buoyant density in CsC1 is 1.51 g cm−3, and S20,w is 410 (data from Mycobacterium phage phlei). Chloroform sensitivity has been reported in possibly related phages.
Nucleic acid
Genomes are about 52 kbp. The Mycobacterium phage L5 (L5) genome has been sequenced and has 52,297 bp. The G+C content is about 63%.
Proteins
Virions contain at least six structural proteins (19–22 to 250 kDa), including a major capsid protein of 35 kDa. Type A (E. coli Pol I) DNA polymerase is encoded.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genetic map is linear. Related genes cluster together. The genome includes 88 genes and has cohesive ends. Host syntheses are shut off during replication. Transcription of structural genes is unidirectional.
Biological properties
Phages are temperate and specific for mycobacteria. Prophages integrate at specific sites in the bacterial genome.
Species demarcation criteria in the genus
Species differ by relative insertions and deletions in the context of otherwise similar sequence and gene organization.
List of species in the genus “L5-like viruses”
| Mycobacterium phage D29 |
|
|
| Mycobacterium phage D29 | [AF022214] | (D29) |
| Mycobacterium phage L5 |
|
|
| Mycobacterium phage L5 | [Z18946] | (L5) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “L5-like viruses” but have not been approved as species
| Mycobacterium phage Bxb1 | [AF271693] | (Bxb1) |
| Mycobacterium phage Leo |
| (Leo) |
| Mycobacterium phage minetti |
| (minetti) |
| Mycobacterium phage phlei |
| (GS4E) |
Genus “c2-like viruses”
Type species Lactococcus phage c2
Distinguishing features
Heads are prolate; phage DNA has cos sites and codes for a putative type B DNA polymerase.
Virion properties
Morphology
Virions have prolate heads about 56×41 nm and tails of 98×9 nm, with a collar (inconstant) and small base plate, and produce rare morphological aberrations (two heads joined by a bridge).
Physicochemical and physical properties
Virion buoyant density in CsC1 is 1.46 g cm−3.
Nucleic acid
Genomes are about 22 kbp (22,163–22,195 bp) and have a G+C content of 35–40%. The genomes of Lactococcus phage c2 (c2) and Lactococcus phage bIL67 (bIL67) have been fully sequenced.
Proteins
Virions have at least six structural proteins (19.2–175 kDa): major proteins are 29, 90 and 175 kDa. Type B (E. coli Pol II) DNA polymerase is apparently encoded.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genetic map is linear. The genome includes 37–38 genes in two clusters and has cohesive ends. Early and late genes are separated by an intergenic region. Late genes are all transcribed in the same direction.
Biological properties
Phages are temperate and specific for lactococci.
Species demarcation criteria in the genus
Phages bIL67 and c2 share 80% nucleotide sequence identity but comparisons also show several DNA deletions or insertions which corresponded to the loss or acquisition of specific ORFs.
List of species in the genus “c2-like viruses”
| Lactococcus phage bIL67 |
|
|
| Lactococcus phage bIL67 | [L33769] | (bIL67) |
| Lactococcus phage c2 |
|
|
| Lactococcus phage c2 | [L48605] | (c2) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “c2-like viruses” but have not been approved as species
| Lactococcus phage c6A | (PBc6A) |
| Lactococcus phage P001 | (P001) |
| Lactococcus phage vML3 | (vML3) |
| (Lactococcus phage ML3) | (ML3) |
| (Lactococcus phage ml3) | (ml3) |
| (Lactococcus phage 3ML) | (3ML) |
About 200 additional, poorly characterized lactococcal phages have been reported that are morphologically indistinguishable from c2.
Genus “PsiM1-like viruses”
Type species Methanobacterium phage psiM1
Distinguishing features
The host is an archaeon. Viral genomes are circularly permuted and terminally redundant.
Virion properties
Morphology
Virions have isometric heads 55 nm in diameter and tails of 210 10 nm with a terminal knob.
Physicochemical and physical properties
Not known.
Nucleic acid
Genomes are about 30 kbp in size.
Proteins
Not known.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
Genomes are circularly permuted and terminally redundant.
Biological properties
Phages are lytic and infect members of the genus Methanobacterium.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus “PsiM1-like viruses”
| Methanobacterium phage psiM1 |
|
|
| Methanobacterium phage ΨM1 | [AF065411, AF065412] | (ΨM1) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “PsiM1-like viruses” but have not been approved as species
| Methanobacterium phage FF3 | (FF3) |
| Methanobrevibacter phage PG | (PG) |
Genus “PhiC31-like viruses”
Type species Streptomyces phage phiC31
Distinguishing features
Phage DNA has cos ends, codes for a type A DNA polymerase and has a serine site-specific recombinase.
Virion properties
Morphology
Virions have isometric heads about 53 nm in diameter and flexible tails 100 nm long and 5 nm wide, a base plate of 15 nm and four tail fibers with terminal knobs (“toes”).
Physicochemical and physical properties
Virion bouyant density is 1.493 g cm−3, and virions are chloroform-sensitive.
Nucleic acid
Genomes are about 43 kbp. Two genomes have been completely sequenced: phages C31 and BT1 are 41,491 and 41,832 bp respectively. G+C content is 63.6%.
Proteins
Virions contain 10 structural proteins, visible by Coomassie staining, of about 10–70 kDa.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genetic map is linear and related genes cluster together. The phage C31 genome encodes 54 genes and has cohesive ends with 10 nt protruding at the 3′ end. Transcription of all except one gene is unidirectional. One tRNA is encoded. The mode of replication is unknown, but the genome encodes a DNA polymerase, phage P4-like primase-helicase, D29-like dCMP deaminase and T4-like nucleotide kinase. Head assembly genes most closely resemble those of Pseudomonas phage D3 and Enterobacteria phage HK97. The putative tail fiber gene contains a collagen motif. Lytic growth occurs via transcription from multiple conserved promoters in the early region and a single operon in the late region. A repressor gene encodes three nested N-terminally different in-frame proteins which bind to multiple highly conserved operators. The integrase belongs to the serine recombinase family of site-specific recombinases.
Biological properties
Phages are temperate and specific for Streptomyces spp. Phages integrate at a specific site in the host genome and are not UV inducible. Phages homoimmune to C31 are susceptible to a phage resistance mechanism (Pgl; phage growth limitation) in S. coelicolor A3(2). Lytic growth switches off host transcription.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus “PhiC31-like viruses”
| Streptomyces phage phiC31 |
|
|
| Streptomyces phage C31 | [AJ006589] | (C31) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “PhiC31-like viruses” but have not been approved as species
| Streptomyces phage BT1 | [AJ550940] | (BT1) |
| Streptomyces phage TG1 |
| (TG1) |
| Streptomyces phage SEA |
| (SEA) |
| Streptomyces phage R4 |
| (R4) |
| Streptomyces phage VP5 |
| (VP5) |
| Streptomyces phage RP2 |
| (RP2) |
| Streptomyces phage RP3 |
| (RP3) |
Genus “N15-like viruses”
Type species Enterobacteria phage N15
Distinguishing features
Prophage DNA is present as a linear plasmid with covalently closed hairpin telomeres. Virion DNA has cohesive ends and is packaged as a unit-size molecule.
Virion properties
Morphology
Phage heads are hexagonal in outline (probable icosahedra), about 60 nm in diameter. Tails are non-contractile, flexible, measure 140×8 nm, and have short brush-like terminal fibers.
Physicochemical and physical properties
Not characterized in detail.
Nucleic acid
The genome is 46,363 bp, has a G+C content of 51.2%, 12 nt 5′-protruding cohesive ends, and is non-permuted. The genome has been sequenced.
Proteins
Virion proteins have not been studied, but their high level of similarity to those of phage λ heads and phage HK97 tails suggests that they are very like those phages.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The virion genome includes about 50 genes and has cohesive ends. Related functions cluster together. The infecting DNA circularizes and replicates, or becomes established as a linear plasmid which is a circular permutation of the virion DNA. Details of DNA replication have not been studied. Organization and sequences of the late expressed virion structural protein and lysis genes are similar to lambda, but the early expressed genes are very different. An anti-repressor system and putative DNA polymerase gene (primase type) have been identified in the early left operon. Unique to this phage type is the presence of a protelomerase gene that encodes an enzyme that resolves a circular genome molecule at the telRL site into the linear molecule with covalently closed hairpin telomeres. An unusually large number of phage genes are expressed from the prophage.
Biological properties
Phages are temperate and mitomycin C inducible. Prophages are linear plasmids with covalently closed hairpin telomeres.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus “N15-like viruses”
| Enterobacteria phage N15 |
|
|
| Enterobacteria phage N15 | [AF064539] | (N15) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “N15-like viruses” but have not been approved as species
| Yersinia phage PY54 | (PY54) |
Genus “SPbeta-like viruses”
Type species Bacillus phage SPbeta
Distinguishing features
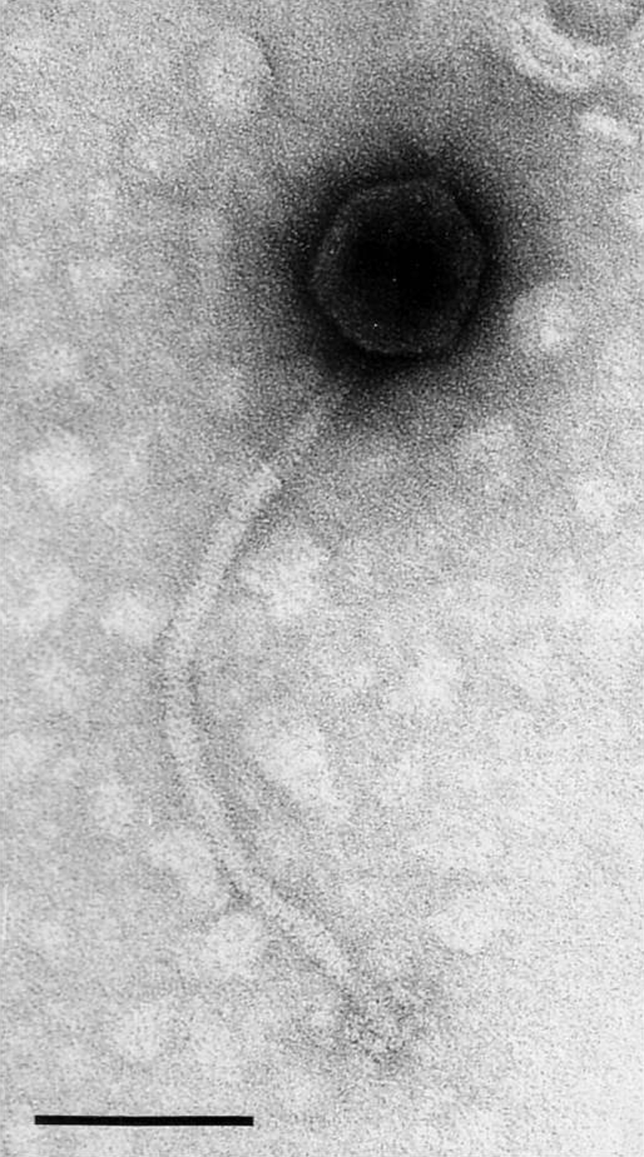
The virions have the largest heads and longest tails in the family Siphoviridae (Figure 3).
Virion properties
Morphology
Heads are icosahedra of 81 nm in diameter. Tails measure 355×10 nm, are relatively rigid, have no collar and possess six club-shaped terminal spikes.
Physicochemical and physical properties
Virion buoyant density in CsCl is 1.52 g cm−3.
Nucleic acid
The genome of SPβ is 134.416 kb in size, consists of 187 ORFs, and has a G+C content of 34–35%. Thymidylate synthetase genes are absent in SPβ but present in other members of the group (3T, 11 isolates).
Proteins
Virions have 6–7 major proteins.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genetic map is linear. Based on the origin of transcription, the genome is divided into three main clusters, the second of which may correspond to late genes. The genome is mosaic and contains bacterial elements, especially in B. subtilis, and elements of eight morphologically unrelated phages or prophages. Only 25% of ORFs have significant homology to known sequences.
Biological properties
These are temperate phages.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus “SPbeta-like viruses”
| Bacillus phage SPbeta |
|
|
| Bacillus phage SPbeta |
| (SPbeta) |
| Bacillus phage SPBc | [AF020713] | (SPBc2) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “SPbeta-like viruses” but have not been approved as species
None reported.
List of unassigned species in the family Siphoviridae
None reported.
Phylogenetic relationships within the family
Refer to the discussion of this topic under Order Caudovirales.
Similarity with other taxa
Refer to the discussion of this topic under Order Caudovirales.
Derivation of name
Sipho: from Greek siphon, “tube”, referring to the long tail.
Further reading
Hatfull, G.F. and Sarkis, G.J. (1993). DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol. Microbiol., 7, 395-405.
Hatfull, G.F., Cresawn, S.G. and Hendrix, R.W. (2008). Comparative genomics of the mycobacteriophages: insights into bacteriophage evolution Res. Microbiol., 159, 332-339.
Lubbers, M.W., Waterfield, N.R., Beresford, T.P.J., Le Page, R.W.F. and Jarvis, A.W. (1995). Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl. Environ. Microbiol., 61, 4348-4356.
Ravin, V., Ravin, N., Casjens, S., Ford, M., Hatfull, G. and Hendrix, R. (2000). Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol., 299, 53-73.
Robert, M.D., Martin, N.L. and Kropinski, A.M. (2004). The genome and proteome of coliphage T1. Virology, 318, 245-266.
Schouler, C., Ehrlich, S.D. and Chopin, M.-C. (1994). Sequence and organization of the lactococcal prolate-headed bIL67 phage genome. Microbiology, 140, 3061-3069.
Smith, M.C.M., Burns, R.N., Wilson, S.E. and Gregory, M.A. (1999). The complete genome sequence of the Streptomyces temperate phage phi C31: evolutionary relationships to other viruses. Nucl. Acids Res., 27, 2145-2155.
Contributed by
Hendrix, R.W., Casjens, S.R. and Lavigne, R.
Figures
Figures
Figure 1 Enterobacteria phage (): (left) representative diagram of a phage particle; (right) electron micrograph of phage particles with negative staining. The bar represents 100 nm.
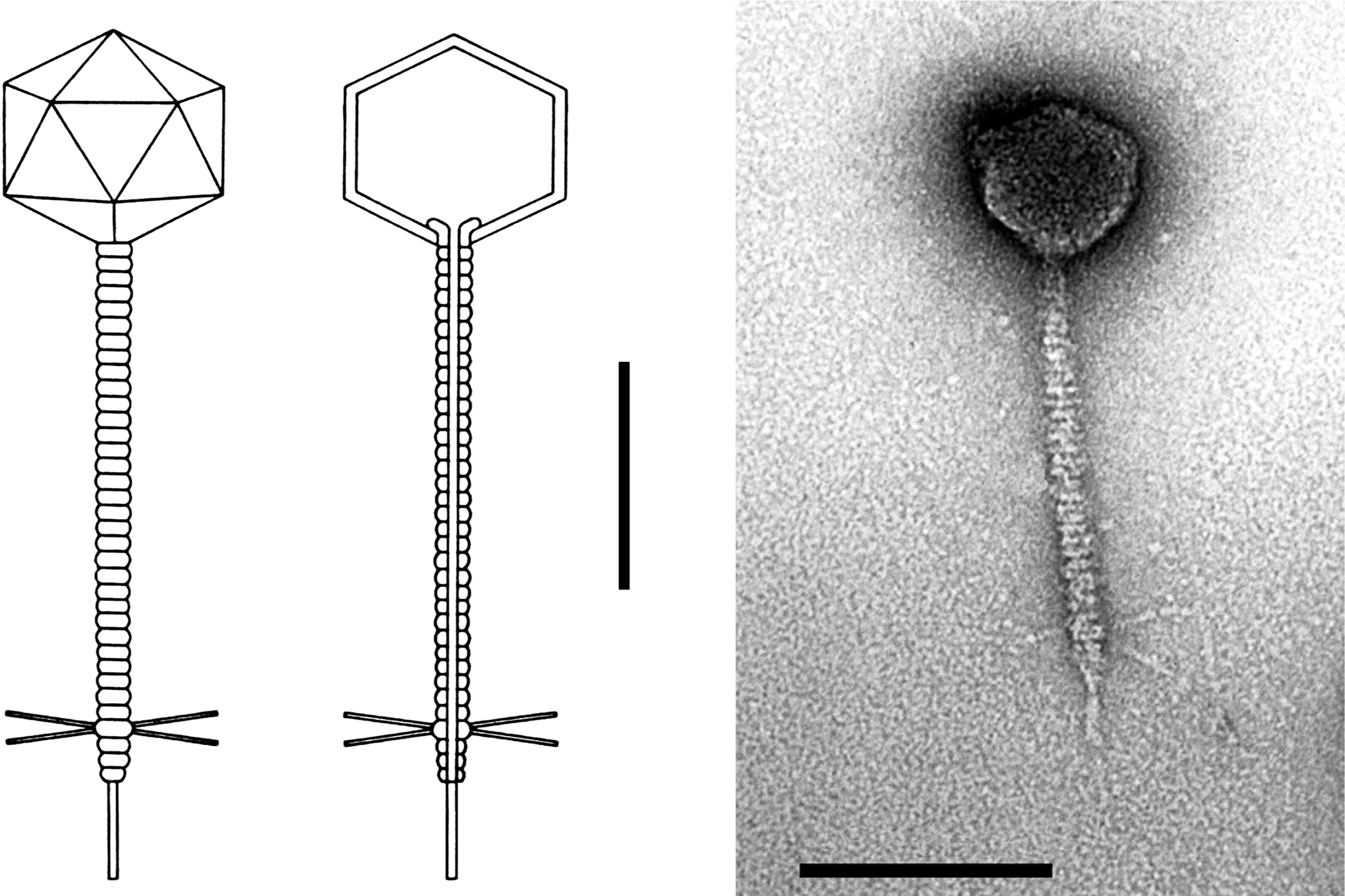
Figure 2 Simplified genetic map of Enterobacteria phage (). Dark portions of the genome indicate non-essential regions.
(From Freifelder, D. (Ed.) (1983). Molecular Biology. Science Books International, Boston, and Van Nostrand Rinehold, New York, p. 639; with permission.)
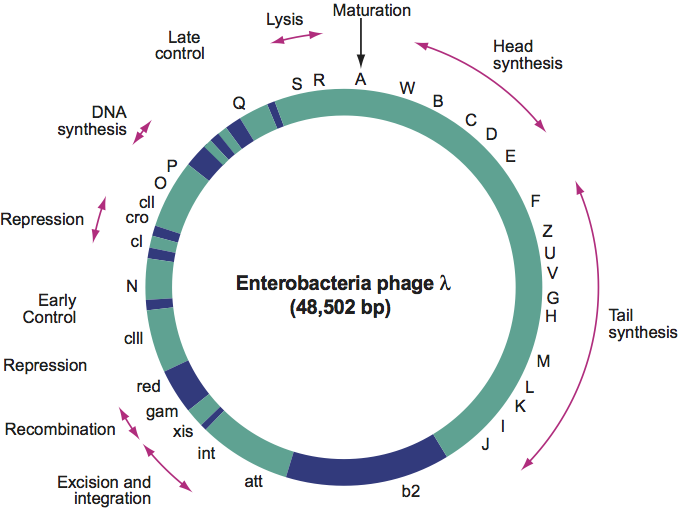
Figure 3 Electron micrograph of Bacillus phage SP particles with negative staining. The bar represents 100 nm.