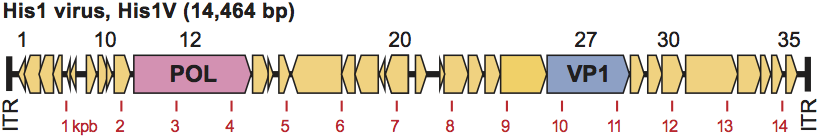Genus: Salterprovirus
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Distinguishing features
Salterproviruses possess a spindle-shaped capsid surrounding a dsDNA linear genome of about 15 kb with inverted terminal repeat sequences and terminal proteins attached at 5′ ends. They replicate via protein-priming and an encoded DNA polymerase. They infect extremely halophilic Archaea.
Virion properties
Morphology
Virus particles are spindle-shaped (44×77 nm) with a short, 7 nm long tail at one end. In Figure 1, many of the particles can be seen attached to host flagella via the tail. Virions can vary somewhat in shape in negative stain preparations, indicating that the capsid is flexible. A small proportion of particles are much larger or more elongated (up to about 204 nm in length). Virions of his2 virus (His2) are even more fragile than those of his1 virus (His1), and negatively-stained preparations usually show severely distorted or disintegrated capsids. However, a few particles can be found that retain the typical spindle shape (44×67 nm).
Physicochemical and physical properties
The buoyant density of His1 in CsCl is 1.28 g cm−3. The virus is sensitive to chloroform, ethanol and detergents such as TritonX-100, but resistant to trichlorotrifluoroethane. Particles are stable if maintained in high salt solutions. Stable over pH range 3–9, and up to 60 °C.
Nucleic acid
The genome of His1 is linear dsDNA, and is 14,464 bp in length. It has 39% G+C, which is significantly lower than that of its host (62.7% G+C). The 5′ ends have protein attached. The related His2 has a genome of similar length, 16,067 bp.
Proteins
The CP(s) or the terminal protein have, as yet, not been determined.
Lipids
Not known.
Carbohydrates
Not known.
Genome organization and replication
His1 has a linear, double stranded DNA genome of 14.5 kb with covalently bound proteins attached to the 5′ termini. The termini contain inverted terminal repeat sequences of 105 bp (Figure 2). The genome is predicted to encode 35 proteins, one of which is a member of the type B group of DNA polymerases, and which uses protein-priming. Replication most likely occurs from both ends of the genome via protein-priming. The His1 genome, with intact terminal proteins, can be transfected into competent cells of Har. hispanica, resulting in the production of infectious virus.
In single-step growth curves, virus release is seen to occur well before significant cell lysis, indicating that virus exit is not linked to lysis. Given the nature of the host cell wall, a thin layer of protein immediately outside of the cell membrane (S-layer), virus particles may bud from the cell, as observed for the morphologically similar Sulfolobus spindle-shaped virus (SSV-1).
Antigenic properties
Not known.
Biological properties
His1 was isolated from a saltern crystallizer pond (Avalon, Victoria, Australia), in which the water was at salt saturation. Not surprisingly, virus particles are most stable at high salt concentration. The isolating host was Haloarcula hispanica, a member of the extremely halophilic Archaea (family Halobacteriaceae). The virus name comes from the first three letters of the host species name. Infection is lytic, but the virus is also capable of existing in an unstable carrier state. Virus particles are able to exit the cell without causing cell lysis. The entire virus genome does not integrate into that of the host, but homologs of the His2 VP1 gene (and 2–3 adjacent genes, probably also membrane proteins) are found in a wide variety of haloarchaeal genome sequences, usually clustered together as a block and keeping the same order and relative orientation. These gene blocks are often nearby genes related to other haloviruses (e.g. HRPV-1, HHPV-1) or haloarchaeal plasmids (e.g. pHK2). Their distribution and significance are currently being studied.
Species demarcation criteria in the genus
Not yet determined.
List of species in the genus Salterprovirus
| His1 virus |
|
|
| His1 virus - DSM22484 | [AF191796] | (His1V-DSM22484)a |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
a Deposited with the German culture collection (DSMZ).
List of other related viruses which may be members of the genus Salterprovirus but have not been approved as species
| His2 virus | [AF191797] | (His2V) |
Phylogenetic relationships within the genus
The genomes sequences of His1 and His2 share little nucleotide similarity but the predicted protein sequences of their DNA polymerase genes share 42% amino acid identity, confirming their specific, phylogenetic relationship.
Similarity with other taxa
Morphologically, virus capsids resemble spindle-shaped archaeal viruses of Sulfolobus (SSV1) but on all other available criteria – sequence similarity, genome structure and replication – they appear to be very different.
Derivation of name
Salterpro: acronym of salt terminal proteins, referring to their characteristic genome structure of linear dsDNA with 5′-attached terminal proteins.
Further reading
Bath, C. and Dyall-Smith, M.L. (1998). His1, an archaeal virus of the Fuselloviridae family that infects Haloarcula hispanica. J. Virol., 72, 9392-9395.
Bath, C., Cukalac, T., Porter, K. and Dyall-Smith, M. (2006) His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus. Virology, 350, 228-239.
Dyall-Smith, M.L., Pfeiffer, F., Klee, K., Palm, P., Gross, K., Schuster, S., Rampp, M. and Oesterhelt, D. (2011). Haloquadratum walsbyi: limited diversity in a global pond. (under review at PLoS ONE).
Porter, K. and Dyall-Smith, M. (2006). Methods for the isolation and study of viruses of halophilic microorganisms. In: Oren, A. and Rainey, F.A. (Eds.), Extremophiles, vol. 35. Elsevier, Amsterdam, pp. 681-702.
Porter, K. and Dyall-Smith, M.L. (2008). Transfection of haloarchaea by the DNAs of spindle and round haloviruses and the use of transposon mutagenesis to identify non-essential regions. Mol. Microbiol. 70, 1236-1245.
Contributed by
Dyall-Smith, M.L.
Figures
Figure 1 Negative-stain electron-micrograph (2% uranyl acetate) of an isolate of His1 virus. Many particles are seen binding to contaminating flagella derived from the host, Haloarcula hispanica. The bar represents 200 nm.
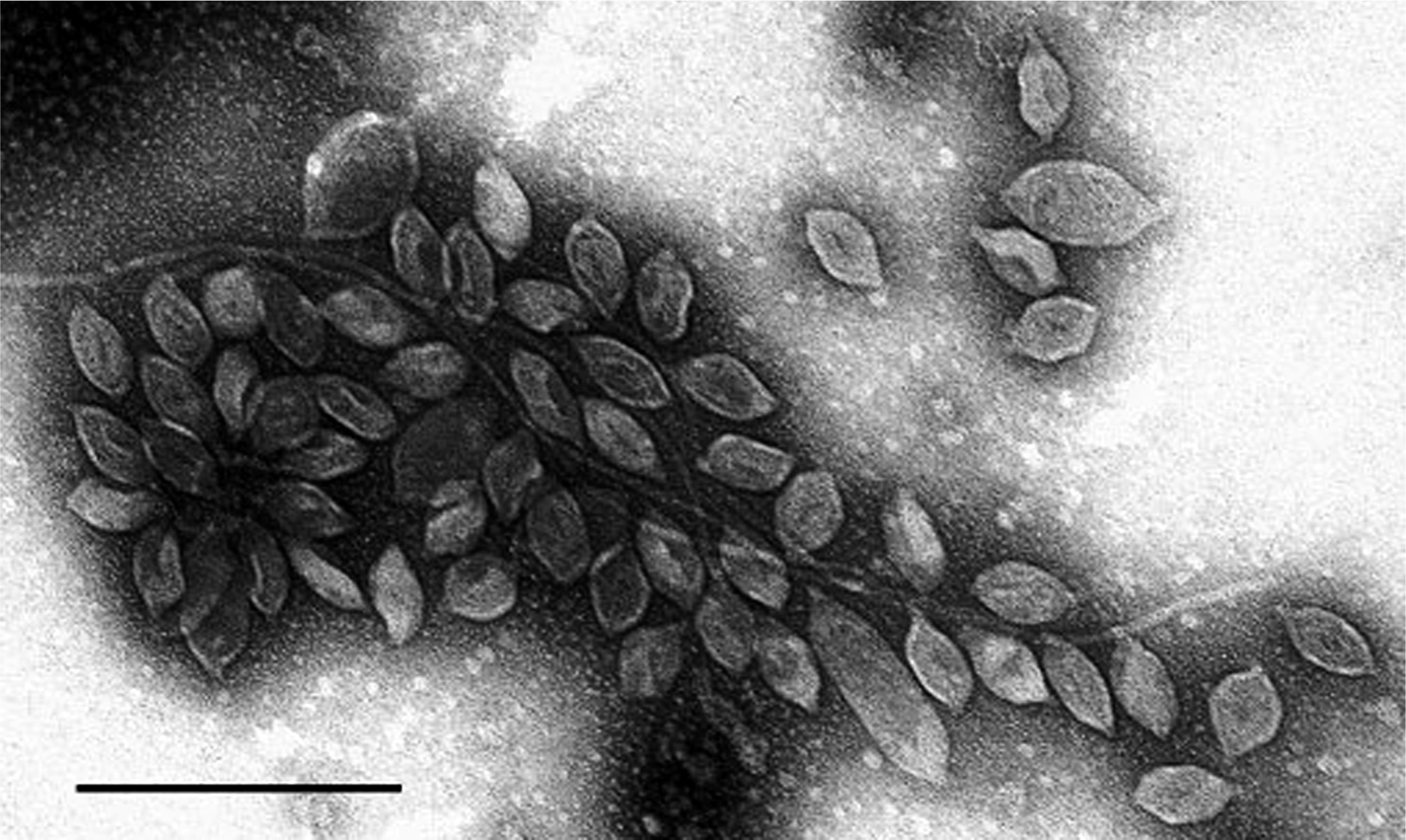
Figure 2 His1 genome, showing predicted ORFs (numbers above), inverted terminal repeats (ITR), the DNA polymerase (POL) and probable major capsid protein gene (VP1). Scale shown underneath is in kbp.