Family: Polyomaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Polyomavirus
Type species Simian virus 40
Virion properties
Morphology
Virions are non-enveloped and approximately 40–45 nm in diameter. The icosahedral capsid is composed of 72 capsomers in a skewed (T=7d) lattice arrangement (Figure 1). Right-handed (dextro) skew has been shown for all polyomaviruses examined in cryoelectron-microscopy tilt experiments. Aberrant structures such as empty capsids, microcapsids and tubular forms are regularly observed.
Physicochemical and physical properties
Virion Mr is 2.5×107. Buoyant density of virions in sucrose and CsCl gradients is 1.20 and 1.34–1.35 g cm−3, respectively. Virion S20,w is 240. Virions are resistant to ether, acid and heat treatment (50 °C, 1 h). Virions are unstable at 50 °C for 1 h in the presence of 1 M MgCl2. Greater than 70% of the total virion protein content is Vp1. Recombinant polyomavirus Vp1 (rVp1), expressed from baculovirus or plasmid constructs in eukaryotic cells, self-assembles into virus-like particles (VLPs) under specific chemical and physical conditions (expression of rVp1 in bacteria leads only to capsomeres). These rVp1-VLPs resemble native virions by electron microscopy and are purified by identical procedures (Figure 2). The rVp1 independently forms pentameric assembly units (pentamers) analogous to the capsomeres of virions, minus the centrally located Vp2 or Vp3 proteins. Linking of the carboxy-termini of pentameric rVp1 creates the icosahedral lattice structure of VLPs. Pentamers that form from rVp1 molecules with truncated carboxy-termini are stable, but do not form VLPs. By modifying the chemical conditions, rVp1-VLPs can be dissociated and subsequently reconstituted. During self-assembly or reconstitution, rVp1-VLPs will non-specifically encapsidate genetic material that is present.
Nucleic acid
Virions contain a single molecule of circular dsDNA. The genomic size is fairly uniform within the genus, averaging approximately 5 kbp. Simian virus 40 (SV40) strain 776 is 5,243 bp, JC polyomavirus (JCPyV) strain Mad-1 is 5,130 bp, BK polyomavirus (BKPyV) strain Dunlop is 5,153 bp, murine polyomavirus (MPyV) strain A2 is 5,297 bp, and baboon polyomavirus 2 (BPyV) is 4,697 bp. The DNA constitutes about 10–13% of the virion by weight. The G+C content varies between 40 and 50%. In the mature virion, the viral DNA is associated with host cell histone proteins H2a, H2b, H3 and H4 in a supercoiled, chromatin-like complex.
Proteins
Currently, polyomavirus genomes are known to code for 5–9 proteins, with sizes predicted from the nucleic acid sequences ranging from 7 to 88 kDa (Table 1). Transcription from one side of the viral origin of DNA replication (ORI) results in mRNAs encoding the early proteins. These non-structural proteins are referred to as tumor (T) antigens because they interfere with cell cycle regulation and, in some cases, induce cellular transformation or tumor formation. Alternative splicing appears to be responsible for the 2–5 related, yet distinct, proteins expressed from each polyomavirus T gene. The set of proteins expressed from a single T gene shares amino-terminal sequence. The T antigens initiate bi-directional viral genome replication, as well as transcription of late viral mRNAs. Late mRNA is transcribed from the strand complementary to that used for early transcription and is also initiated from the opposite side of the ORI. Late transcripts code for three structural proteins (Vp1, Vp2 and Vp3) as well as another non-structural protein known as agnoprotein. Of the three structural proteins, Vp1 makes up more than 70% of the total virion protein content and hence is also referred to as the major structural protein. Five Vp1 proteins surround either a Vp2 or Vp3 molecule to form stable assembly units, or capsomers; 72 random capsomers link together in icosahedral symmetry to form the capsid of each virion. The Vp2 and Vp3 molecules may be necessary to ensure specific encapsidation of the replicated polyomavirus genome. Also, VP2 is myristylated (at least in MPyV and SV40) and has a possible role in entry. The agnoprotein may have some role in facilitating capsid assembly, but it is not a component of the mature virion.
The following recently discovered polyomavirus-coded proteins are not listed in Table 1. The SV40 late region, and perhaps that of other mammalian polyomaviruses as well, encodes a 15 kD protein called Vp4 that is not contained in capsids. In the case of SV40, Vp4 appears to be involved in host cell lysis. The recently discovered Merkel cell polyomavirus (MCPyV) encodes a splice variant of the SV40 17kT protein. Except for the recently-discovered canary polyomavirus (CaPyV) (Ref: PMID: 20797969), known avian polyomaviruses have an additional ORF in the late region, at a site corresponding to that of the agnoprotein ORF in mammalian polyomavirus genomes. The protein encoded by this avian polyomavirus ORF is designated Vp4 (also referred to as agnoprotein 1a), but it has no apparent sequence homology to the SV40 Vp4 protein or to the mammalian polyomavirus agnoprotein. Vp4 of avian polyomavirus (APyV; also known as “budgerigar fledgling disease polyomavirus”) is a component of the mature virion. It is thought to be involved in packaging the viral genome and inducing apoptosis. Other APyV-coded proteins not listed or designated as such in Table 1 are agnoprotein 1b (same as VP4 delta), agnoprotein 2a and agnoprotein 2b (two poorly characterized hydrophobic proteins). Agnoproteins 1a/1b and 2a/2b could either be seen as functional proteins or alternatively as non-functional splicing variants. The genomes of KI polyomavirus (KIPyV), WU polyomavirus (WUPyV), human polyomavirus-6 (HPyV6), HPyV7 and MCPyV do not appear to contain ORFs that might encode an agnoprotein, nor do they appear to encode a middle T antigen.
Table 1 Deduced size of polyomavirus proteins
| Virus | MPyV | SV40 | JCPyV | BKPyV | KPyV | LPyV | BPyV | APyV |
| Structural proteins |
|
|
|
|
|
|
|
|
| VP1 | 42.4 | 39.9 | 39.6 (40) | 40.1 (40) | 41.7 | 40.2 | 40.5 | 37.4 (42) |
| VP2 | 34.8 | 38.5 | 37.4 | 38.3 | 37.4 | 39.3 | 39.1 | 37.3 (39) |
| VP3 | 22.9 | 27.0 | 25.7 | 26.7 | 25.2 | 27.3 | 26.9 | 27.0 (30) |
| Non-structural proteins |
|
|
|
|
|
|
|
|
| T | 88.0 | 81.6 (94) | 79.3 (94) | 80.5 | 72.3 | 79.9 | 66.9 | 68.3 (80) |
| mT | 48.6 | ND | ND | ND | ND | ND | ND | ND |
| T′135 | ND | ND | (17) | * | ND | ND | ND | ND |
| T′136 | ND | ND | (17) | * | ND | ND | ND | ND |
| T′165 | ND | ND | (22–23) | * | ND | ND | ND | ND |
| 17kT | ND | (17) | ND | 17–20 | ND | ND | ND | ND |
| tT | * | ND | (17) | ND | ND | ND | ND | ND |
| t | 22.8 | 20.4 | 20.2 | 20.5 | 18.8 | 22.2 | 14.0 | 17.0 (24) |
| LP1/agno | ND | 7.3 | 8.1 | 7.4 | ND | ND | 13.1 | ND |
Predicted sizes of polyomavirus proteins in kDa: ( ), observed sizes of the expressed proteins; *, reported proteins that lack both predicted and expressed size; ND, not detected.
Lipids
None present.
Carbohydrates
None present
Genome organization and replication
Virions that attach to cellular receptors are taken up by the cell and transported to the nucleus, where the genome is transcribed and replicated. During a productive infection, transcription of the viral genome is divided into an early and a late stage. Transcription of the early and late coding regions is controlled by separate promoters through the binding of specific transcription factors and cis-acting elements. The sequence that codes for early transcripts is exclusive to one strand of the viral DNA and spans approximately half the genome. Late transcripts are generated in the opposite direction from the other half of the complementary strand (Figure 3).
Precursor mRNAs undergo post-transcriptional processing, which includes capping and polyadenylation of the 5′ and 3′ termini, respectively, as well as splicing. Efficient use of coding information involves differential splicing of the transcripts and use of overlapping ORFs. Early mRNAs encode regulatory, non-structural proteins that may exhibit cis- or trans-activating properties. These include proteins that are required for initiation of viral DNA replication and late protein production. Their expression leads to de-repression of some host cell enzymes and stimulation of cellular DNA synthesis. Prior to the start of the late events, viral DNA replication is initiated in the nucleus. Translation of most of the late transcripts produces structural proteins that are involved in capsid assembly. Post-translational modifications of some early and late viral proteins include phosphorylation, N-acetylation, fatty acid acylation, ADP-ribosylation, methylamination, adenylation, glycosylation and sulphation. Several of the viral proteins contain nuclear localization signals, which facilitate transport of the proteins to the host cell nucleus, where virion maturation occurs. Virions are released by lysis of infected cells.
Identified non-structural proteins include: large T, middle (m) T and small t for mouse and hamster polyomaviruses; large T, 17kT and small t for SV40. JCPyV also encodes small t, and BKPyV makes the equivalent of the SV40 17kT. In addition to large T and small t, three other large T intermediates, termed T Prime (T′135, T′136 and T′165) have been described for JCPyV. Similar T′ antigens expressed from BKPyV have been identified, but the precise size of these proteins has not been reported. No mRNA encoding a protein of size comparable to the small t proteins of other polyomaviruses has been identified in BKPyV (Table 1). T antigens, first named for their involvement in tumorigenicity and transformation, play key roles in the regulation of transcription and DNA replication. The best characterized of these, SV40 large T antigen, exhibits multiple functions that can be mapped to discrete domains.
Replication of the viral genome is initiated by the specific binding of T antigen to ORI and its interaction with host DNA polymerase(s). Due to the limited amount of genetic information encoded by the viral genomes, the polyomaviruses rely heavily upon host cell machinery, including nuclear transcription factors, to replicate their DNA. Replication proceeds bi-directionally via a “Cairns” structure and terminates about 180° from ORI. Late in the replication cycle, rolling circle-type molecules have been identified. The viral proteins involved in initiation may also promote elongation through helicase and ATPase activities.
The non-coding regulatory region of each polyomavirus is positioned between the early and late protein-coding sequences. This sequence contains promoter/enhancer elements. Within each polyomavirus species, the nucleotide sequence of the regulatory region is hypervariable. Nucleotide sequencing studies have uncovered numerous variations of regulatory region structure. For the human polyomavirus JCPyV, as with other polyomaviruses, the nucleotide sequence of the regulatory region has been shown to control levels of viral transcription and replication. The JCPyV “archetype” regulatory region sequence, which conveys relatively inefficient viral activity, contains a single copy of all sequence sections observed in all other variant forms (Figure 4). From the early side of the archetype, the initial regulatory region sequence section contains ORI followed by sequence sections designated a, b, c, d, e and f. From variant to variant, sequence sections a through e are the most likely to present deletions, replications and/or unique arrangements; for example, deletion of b and d leaves ace, a 98 bp sequence unit. Although the ace sequence unit conveys more activity than archetype, it appears to be the minimal sequence unit required for function. Also, tandem ace sequence units, or repeats, constitute the regulatory region of the more robust “prototype” JCPyV sequence, Mad-1. Such modification to the regulatory region structure appears to alter the cellular host range and may also be responsible for switching JCPyV between states of lytic and latent infection. Arranging all known variant JCPyV regulatory regions into quadrants, according to integration of unique sequence sections and/or repetition of sequence section groups, also links variants by activity. Four distinct structural forms (I-S, I-R, II-S and II-R) are defined along with tissue tropisms. This design, known as the JCV Compass (Figure 4), provides logical connections between variant regulatory regions and may be useful for elucidating crucial steps in JCPyV pathogenesis. Currently, it is not known whether similar arrangements of other polyomavirus species variants also render logical relationships. The consensus sequence of the regulatory region responsible for binding of large T antigen, including that at ORI, is distinctly different in the avian polyomaviruses from that in the mammalian polyomaviruses.
An SV40 DNA sequence, located in the untranslated region 3′ to the polyadenylation cleavage site of the late pre-mRNA, encodes a miRNA that accumulates at late times in infection. This miRNA marks early viral mRNAs for degradation, thereby down-regulating T-antigen expression at late times in infection. This miRNA makes SV40-infected cells less susceptible to attack by cytotoxic T cells in vitro. However, a MPyV variant that cannot produce its miRNA shows no difference in pathogenicity. It is not known whether other polyomaviruses encode miRNAs.
Antigenic properties
The human polyomaviruses JCPyV and BKPyV can be detected by hemagglutination of human type O erythrocytes, while the murine polyomavirus (MPyV) and the goose hemorrhagic polyomavirus (GHPyV) can hemagglutinate sheep and chicken erythrocytes, respectively. The capsids bind to the surface of the erythrocytes, resulting in a three-dimensional lattice-like suspension known as hemagglutination. Using serial dilutions, a titer expressed as hemagglutination units (HA units) can be determined.
Antisera prepared against disrupted virions can also detect antigens shared with other species in the genus. Members of the polyomavirus species can be distinguished antigenically by neutralization, hemagglutination inhibition and immuno-electron microscopy tests. Serum levels of antibodies to JCPyV, BKPyV and SV40 can also be detected by enzyme-linked immunosorbent assay (ELISA) by coating microtiter plates with either whole virion or recombinant VLPs. ELISA tests are also available, and are used routinely for APyV and GHPyV. Polyclonal and monoclonal antibodies can be used to demonstrate cross-reactivity between the T proteins of the primate polyomaviruses. However, there are also specific antibodies currently available that can distinguish among the T antigen epitopes of JCPyV, BKPyV and SV40.
Biological properties
Mammalian polyomaviruses
Whereas each of the mammalian polyomaviruses grows most efficiently in vitro in cells of its natural host, host species-specificity is not absolute. Cells that fail to support viral replication may be transformed by the action of the viral early gene products.
Mammalian polyomaviruses give rise to primary infections in their natural hosts that are usually not associated with clinical syndromes, although BKPyV infections have sometimes been associated with mild urinary tract and upper respiratory symptoms; the latter consistent with a possible respiratory route of BKPyV transmission. Primary infections in natural hosts then generally lead to clinically uneventful, persistent infections. The murine pneumotropic polyomavirus (MPtV) is a notable exception, being able to cause severe acute disease in newborn mice.
The recognized human polyomaviruses, JCPyV and BKPyV, are distributed worldwide, as demonstrated by detectable levels of circulating antibodies in the majority of the healthy human population. These viruses generally establish persistent infections, usually early in life, after which they can remain latent in several body compartments, including the tonsils, lower urinary tract, lymphoid tissues and bone marrow. Involvement of the kidney is frequently observed, with viruria noted, especially in immunodeficient hosts and patients undergoing renal transplantation.
The exact routes of mammalian polyomavirus transmission are unclear. Since SV40, JCPyV and BKPyV are each known to target the urinary tract, low-level shedding of virus in urine is thought to play a role in transmission of these polyomaviruses. However, JCPyV and BKPyV have each been found in tonsillar tissue, consistent with respiratory transmission or transmission by hand to mouth contact. Virus spread may also occur when persistent infections are reactivated during periods of immune suppression, including pregnancy. Transmission via tissue transplantation is also thought to play a role in humans. Vectors are not known to play a role in transmission of polyomaviruses.
Whereas BKPyV and JCPyV infections are generally asymptomatic or associated with mild pathologic changes in the respiratory and urinary tracts of immunocompetent individuals, BKPyV infection is an increasingly common complication in transplant recipients, resulting in nephropathy or cystitis; a significant cause of kidney transplant failure. Moreover, there have been reports of disseminated BKPyV infections that gave rise to meningitis, retinitis, pneumonia or vasculopathy. In severely immunocompromised individuals, JCPyV can infect and destroy oligodendrocytes of the central nervous system, thereby giving rise to a fatal demyelinating disease termed progressive multifocal leucencephalopathy (PML). PML is a common complication in HIV-1 infection, eventually affecting 5% of the AIDS population. There is no effective treatment for PML, but the introduction of highly active antiretroviral therapy (HAART) has resulted in a significant decline in the HIV/PML mortality rate. SV40 may cause a PML-like disease in rhesus monkeys, particularly in those infected with HIV.
Two recently discovered putative (insofar as they have not yet been cultivated by inoculating cells in culture) human polyomaviruses, KIPyV and WUPyV, were initially detected in nasopharyngeal aspirates from patients presenting with acute respiratory tract infections, but it is not yet clear whether these viruses are agents of human respiratory tract disease. The association of integrated DNA of the recently discovered MCPyV with human Merkel cell carcinomas, as confirmed by several research groups, may represent the first human malignancy associated with the consistent presence of a particular polyomavirus genome. Interestingly, the MCPyV genomic sequence that encodes the large T antigen was shown to contain a chain-terminating mutation in 9/9 Merkel cell carcinomas. A similar mutation was not seen in MCPyV genomes from non-tumor sources. These experimental findings are consistent with the premise that MCPyV genomes in tumors undergo T-antigen mutations that prevent integrated virus replication (which would lead to cell death), while not effecting oncogenesis. Nevertheless, the role of MCPyV in Merkel cell carcinoma remains uncertain.
Using a rolling circle amplification technique, the genomes of two novel, currently unclassified polyomaviruses, human polyomavirus 6 (HPyV6) and human polyomavirus 7 (HPyV7), were detected in skin swabs from healthy human adults. They are closely related to each other and to WUPyV and KIPyV, but are distantly related to MCPyV. Genomes of the latter virus were also detected in these skin samples, suggesting that at least three polyomaviruses species may be commonly present in human skin.
Beta-lymphotropic polyomavirus (LPyV, or AGMPyV; see list of species in the genus) was isolated from a lymphoblastoid cell line derived from an African green monkey. Nevertheless, LPyV can productively infect some human B cell lymphoma-derived cell lines. This tropism is noteworthy because seroprevalence data suggest that a serologically related counterpart to LPyV circulates in humans, with a prevalence level of 15–20% in adults. Although phylogenetic analyses suggest that LPyV is closely related to MCPyV (in a cluster distinct from that containing WUPyV and KIPyV (Figure 5), the serology of the monkey and putative human viruses is distinct. LPyV transforms hamster embryo cells in vitro and induces choroid plexus tumors in transgenic mice that express the LPyV early region. The nature of the supposed human counterpart to LPyV and its possible role in human disease are not yet clear.
At present, there is increasing interest in the medical importance of the human polyomaviruses. The reasons are as follows: (1) polyomaviruses are ubiquitous in humans, (2) polyomaviral pathology is usually seen only in immunocompromised individuals, and (3) the frequency of immmunocompromised individuals has been on the rise because of the increasing numbers of the elderly, tissue transplantation, therapeutic immunomodulators and AIDS.
In November 2009, using a broad-spectrum PCR assay, two new, distinctly different provisional polyomaviruses were reported; one in Bornean orangutans (OranPyV-1) and the other in Sumatran orangutans (OranPyV-2). Sequencing showed that the genomes of each of these putative polyomaviruses has the characteristic genome architecture, but lacks an obvious agnogene. In addition, using PCR primers designed to identify polyomavirus genomes, putative polyomaviruses were identified in two species of bat (Myotis lucifugus and M. californicus). These findings may be medically relevant, considering that known instances of emerging diseases in humans are caused by viruses endemic to bats that have been transmitted to humans either directly or through other animal intermediaries.
MPtV is somewhat unique among the mammalian polyomaviruses, with respect to being able to cause severe disease during primary infection. In newborn mice, MPtV causes highly lethal interstitial pneumonia, perhaps explained by its ability to replicate in vascular endothelial cells of the lung. However, infection generally leads to asymptomatic, persistent infection in immunocompetent adult animals.
Most mammalian polyomaviruses induce neoplastic transformation in cell culture and tumors in rodents and some primates. Transformation in vitro and tumorigenesis in vivo result from expression of virus-coded early proteins, which interact with specific cellular proteins (p53, pRB and others). Polyomavirus genomes are usually integrated into chromosomes of transformed or tumor cells. In vivo, JCPyV can induce brain tumors in owl and squirrel monkeys, and there has been recent interest in the possibility that JCPyV infection might lead to the development of central nervous system tumors in humans. However, this issue remains controversial. As noted above, MCPyV has been associated with Merkel cell carcinomas, although the role of the virus in this neoplasm is as yet uncertain. The inadvertent exposure of millions of human poliovirus vaccine recipients to SV40 (a previously unrecognized contaminant of early vaccines) led to concern that this virus may be a cause of human neoplasms and that it may yet be circulating in the human population. These issues also remain controversial. MPyV produces a wide variety of tumors in its natural host.
Avian polyomaviruses
The biology of the recognized avian polyomaviruses (ApyVs) is markedly distinct from that of the mammalian polyomaviruses in several key respects. First, avian viruses display a high degree of pathogenicity, leading to acute and chronic inflammatory diseases, especially in young birds. Second, none of the avian polyomaviruses displays the ability to induce neoplasia. Third, avian polyomavirus displays a broad host range, in comparison to the rather restricted host ranges of the mammalian polyomaviruses. Indeed, whereas the ICTV assigned the species name Budgerigar fledgling disease virus in recognition of its typical disease pattern in budgerigars, it is now referred to as APyV in recognition of its broad host range. Moreover, whereas the mammalian polyomaviruses typically display a distinct tissue tropism, APyV is able to replicate in a wide variety of organs. Likewise, GHPyV, the etiological agent of hemorrhagic nephritis and enteritis of geese, displays a broad tissue tropism in geese. A high seroprevalence rate was seen in Germany even in asymptomatic geese. Neither the tissue tropisms nor clinical importance of Finch polyomavirus and Crow polyomavirus have been well characterized.
Species demarcation criteria in the genus
Species and genus demarcation criteria are currently being developed. In the interim, the list of species is provisional, and all species are assigned to a single genus.
List of species in the genus Polyomavirus
| African green monkey polyomavirus |
|
|
| African green monkey polyomavirus | [K02562] | (AGMPyV) |
| (Beta-lymphotropic polyomavirus) |
| (LPyV) |
| Baboon polyomavirus 2 |
|
|
| Baboon polyomavirus 2 |
| (BPyV-2) |
| BK polyomavirus |
|
|
| BK polyomavirus | [V01108] | (BKPyV) |
| Bovine polyomavirus |
|
|
| Bovine polyomavirus | [D13942=NC_001442] | (BPyV) |
| Budgerigar fledgling disease polyomavirus |
|
|
| Avian polyomavirus | [AF241168] | (APyV) |
| Hamster polyomavirus |
|
|
| Hamster polyomavirus | [AJ006015=NC_001663] | (HaPyV) |
| Human polyomavirus |
|
|
| Human polyomavirus |
|
|
| JC polyomavirus |
|
|
| JC polyomavirus | [J02226=NC_001699] | (JCPyV) |
| Murine pneumotropic virus |
|
|
| Murine pneumotropic virus | [M55904] | (MPtV) |
| Murine polyomavirus |
|
|
| Murine polyomavirus | [J02288] | (MPyV) |
| Rabbit kidney vacuolating virus |
|
|
| Rabbit kidney vacuolating virus |
| (RKV) |
| Simian virus 12 |
|
|
| Simian virus 12 | [AY614708] | (SV12) |
| (SA12) |
|
|
| (Baboon polyomavirus type 1) |
|
|
| Simian virus 40 |
|
|
| Simian virus 40 | [J02400=NC_001669] | (SV40) |
Species names are in italic script; names of isolates are in roman script; synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Polyomavirus but have not been approved as species
| Mammalian viruses |
|
|
| KI polyomavirus | [EF127906] | (KIPyV) |
| WU polyomavirus | [EF444549] | (WUPyV) |
| Merkel cell polyomavirus | [EU375803] | (MCPyV) |
| Canary polyomavirus | [GU345044] | (CaPyV) |
| Cynomolgus polyomavirus |
| (CyPV) |
| Chimpanzee polyomavirus | [AY691168] | (ChPyV) |
| Athymic rat polyomavirus |
| (Rat-PyV) |
| Squirrel monkey polyomavirus | [NC_009951] | (SqPyV) |
| Bornean orangutan polyomavirus | [FN356900] | (OraPyV-1) |
| Sumatran orangutan polyomavirus | [FN356901] | (OraPyV-2) |
| Bat polyomavirus | [NC_011310] | (BatPyV) |
| Human polyomavirus 6 | [NC_014406] | (HPyV6) |
| Human polyomavirus 6 | [NC_014407] | (HPyV7) |
| Avian viruses |
|
|
| Crow polyomavirus | [NC_007922] | (CPyV) |
| Finch polyomavirus | [NC_007923] | (FPyV) |
| Goose hemorrhagic polyomavirus | [NC_004800] | (GHPyV) |
These viruses have not been approved as species because, although distinctive polyomaviral genomes have been detected by PCR-based screening procedures, viruses have not yet been cultivated by inoculating cells in culture.
List of unassigned species in the family Polyomaviridae
None reported.
Phylogenetic relationships within the family
Phylogenetic relationships among the polyomaviruses, based on the amino acid sequences of VP1, VP2 and large T antigen, are shown in Figure 5.
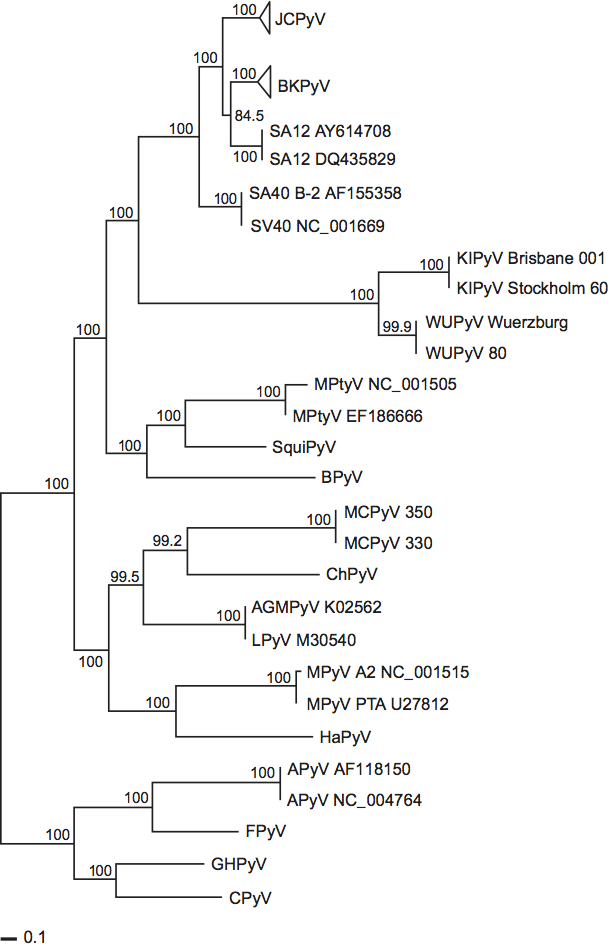
Genetic distances between species are consistent with separate branching of the mammalian and avian polyomaviruses. Moreover, this separate branching is in accord with the premise that polyomaviruses have co-evolved with their natural hosts, as supported by a 2006 analysis of 72 complete genomes. (However, the putative “human” polyomaviruses, KIPyV and WUPyV, do not cluster with established human polyomaviruses, JCPyV and BKPyV, and MCPyV is even more distant in the phylogenetic tree [Figure 5].) Concerning differences in genomic structures, four of the five avian polyomaviruses contain an additional ORF that is created by alternative splicing in the 5′ end of the late coding region. The encoded protein, designated VP4 in APyV, is incorporated into the APyV capsid. It has no counterpart in the mammalian polyomaviruses and it shares no homology with any mammalian polyomavirus protein. In addition, the DNA-binding domain of the mammalian polyomavirus large T antigens have a different consensus sequence from that of the avian polyomaviruses. Specifically, the mammalian polyomaviruses may all use the pentanucleotide GAGGC as the large T antigen-binding sequence, whereas the avian polyomaviruses may use the palindromic motif CC(A/T6)GG. Concerning biological properties, the mammalian and avian polyomaviruses differ as follows. Whereas mammalian polyomaviruses generally give rise to subclinical infections in their immunocompetent natural hosts and may be tumorigenic in laboratory animals, avian polyomavirus infections are associated with severe pathologies, including hepatitis, nephritis and feather disorders, and they are not known to induce tumors. Moreover, at least one of the avian polyomaviruses (APyV) has a notably broader host range than any known mammalian polyomavirus.
Based on phylogenetic data (Figure 5), genomic structure and biological properties, the Polyomaviridae Study Group is investigating the merits of dividing the family into two genera; one containing the mammalian polyomaviruses and a genus containing the avian polyomaviruses. In addition, the Study Group is considering the merits of creating a separate mammalian genus, comprised of KIPyV, WUPyV, HPyV6 and HPyV7, to reflect their nucleotide sequence divergence from the other mammalian polyomaviruses.
Similarity with other taxa
Until the VIIth ICTV report, the genus Polyomavirus was assigned as one of two genera within the family Papovaviridae (the other genus being Papillomavirus). However, sequence data established unequivocally that the Polyomaviridae and Papillomaviridae are not detectably related and constitute distinct virus families.
Bandicoot papillomatosis carcinomatosis virus types 1 and 2 (BPCV1 and BPCV2, respectively) have circular dsDNA genomes that are similar to members of the family Papillomaviridae in size (ca. 7.3–7.5 kbp) and possibly in some aspects of gene content in that they encode putative papillomaviral L1 and L2 structural proteins. However, they also encode putative polyomaviral large T and small t antigens. The origins of these viruses is not clear, but they might be explained by recombination between a polyomavirus and a papillomavirus. Although these as yet unclassified viruses appear not to be true polyomaviruses, they have been listed here for the sake of completeness.
Derivation of name
Polyoma: from Greek poly, “many”, and -oma, denoting “tumors”.
Further reading
Allander, T., Andreasson, K., Gupta, S., Bjerkner, A., Bogdanovic, G., Persson, M.A., Dalianis, T., Ramqvist, T. and Andersson, B. (2007). Identification of a third human polyomvirus. J. Virol., 81, 4130-4136.
Ciappi, S., Azzi, A., De Santis, R., Leoncini, F., Sterrantino, G., Mazzotta, F., Mecocci, L. (1999). Archetypal and rearranged sequences of human polyomavirus JC transcription control region in peripheral blood leukocytes and in cerebrospinal fluid. J. Gen. Virol., 80, 1017-1023.
Dalianis, T., Ramqvist, T., Andreasson, K., Kean, J.M. and Garcea, R.L. (2009). KI, WU and Merkel cell polyomaviruses: a new era for human polyomavirus research. Semin. Cancer Biol., 19, 270-275.
Daniels, R., Sadowicz, D. and Hebert, D.N. (2007). A very late viral protein triggers the lytic release of SV40. PLoS Pathog. 3, e98.
Gaynor, A.M., Nissen, M.D., Whiley, D.M., Mackay, I.M., Lambert, S.B., Wu, G., Brennan, D.C., Storch, G.A., Sloots, T.P. and Wang, D. (2007). Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 3, e64.
Houff, S.A., Major, E.O., Katz, D.A., Kufta, C.V., Sever, J.L., Pittalunga, S., Roberts, J.R., Gitt, J., Saini, N. and Lux, W. (1988). Involvement of JC virus-infected mononuclear cells from the bone marrow and spleen in the pathogenesis of progressive multifocal leukoencephalopathy. N. Engl. J. Med., 318, 301-305.
Imperiale, M.J. and Major, E.O. (2007). Polyomaviruses. In: D.M. Knipe, P.M. Howley, D.E. Griffen, R.A. Lamb, R.A. Martin, B. Roizman and S.E. Straus (Eds.), Fields Virology, 4th edn. Lippincott Williams and Wilkins, Philadelphia, PA, pp. 2263-2298.
Johne, R. and Müller, H. (2007) Polyomaviruses of birds: etiologic agents of inflammatory diseases in a tumor virus family. J. Virol. 81, 11554-11559.
Krumbholz, A., Bininda-Emonds, O.R., Wutzler, P. and Zell, R. (2009). Phylogenetics, evolution, and medical importance of polyomaviruses. Infect. Genet. Evol. 5, 784-799.
Monaco, M.C.G., Jensen, P.N., Hou, J., Durham, L.C. and Major, E.O. (1998). Detection of JC virus DNA in human tonsil tissue: Evidence for site of initial viral infection. J. Virol., 72, 9918-9923.
Pérez-Losada, M., Christensen, R.G., McClellan, D.A., Adams, B.J., Viscidi, R.P., Demma, J.C. and Crandall, K.A. (2006) Comparing phylogenetic codivergence between polyomaviruses and their hosts. J. Virol., 80, 5663-5669.
Schowalter, R.M., Pastrana, D.V., Pumphrey, K.A., Moyer, A.L. and Buck, C.B. (2010). Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host & Microbe, 7, 509-515.
Shuda, M., Feng, H., Kwun, H.J., Rosen, S.T., Gjoerup, O., Moore, P.S. and Chang Y. (2008) T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl Acad. Sci., USA, 105, 16272-16277.
Contributed by
Norkin, L.C., Allander, T., Atwood, W.J., Buck, C.B., Garcea, R.L., Imperiale, M.J., Johne, R., Major, E.O., Pipas, J.M. and Ramqvist, T.
Figures
Figure 1 (Left) Computer rendering of a particle of murine polyomavirus strain A2. (Center) Capsomer bonding relations. Each icosahedral asymmetric unit comprises six Vp1 subunits, including one (a) from a pentavalent pentamer. The six symmetrically different subunits are designated a, a, a, b, b and c, corresponding to three different bonding states. (Right) Computer graphics representation of the surface of the capsid of murine polyomavirus strain A2. Five Vp1 subunits form the basis of a polyomavirus capsomer, and 72 capsomers link together in a 12 pentavalent/60 hexavalent arrangement, conveying icosahedral capsid structure.

(Left and center, from Salunke, D.M., Casper, D.L.D. and Garcea, R.L. (1986). Cell46, 895904; right, from Eckhart W. (1991). In Fundamental Virology, 2nd edn (B.N. Fields and D.M. Knipe, Eds.), Raven Press, New York; with permission.)
Figure 2 Electron micrographs of: (left) brain tissue from a progressive multifocal leukoencephalopathy (PML) patient showing the assembly of JC polyomavirus (JCPyV) particles in the nucleus of an infected oligodendrocyte; and (right) composite of virus-like particles (VLP) self-assembled from recombinant Vp1 of BK polyomavirus (BKPyV) (rBKVp1), purified by CsCl ultracentrifugation from supernatants of Sf9 insect cell cultures infected with recombinant BKVp1-baculovirus. The particles were negatively stained with 2% phosphotungstic acid. The composite includes an insert showing an enlargement of a single VLP. Bar=100 nm.
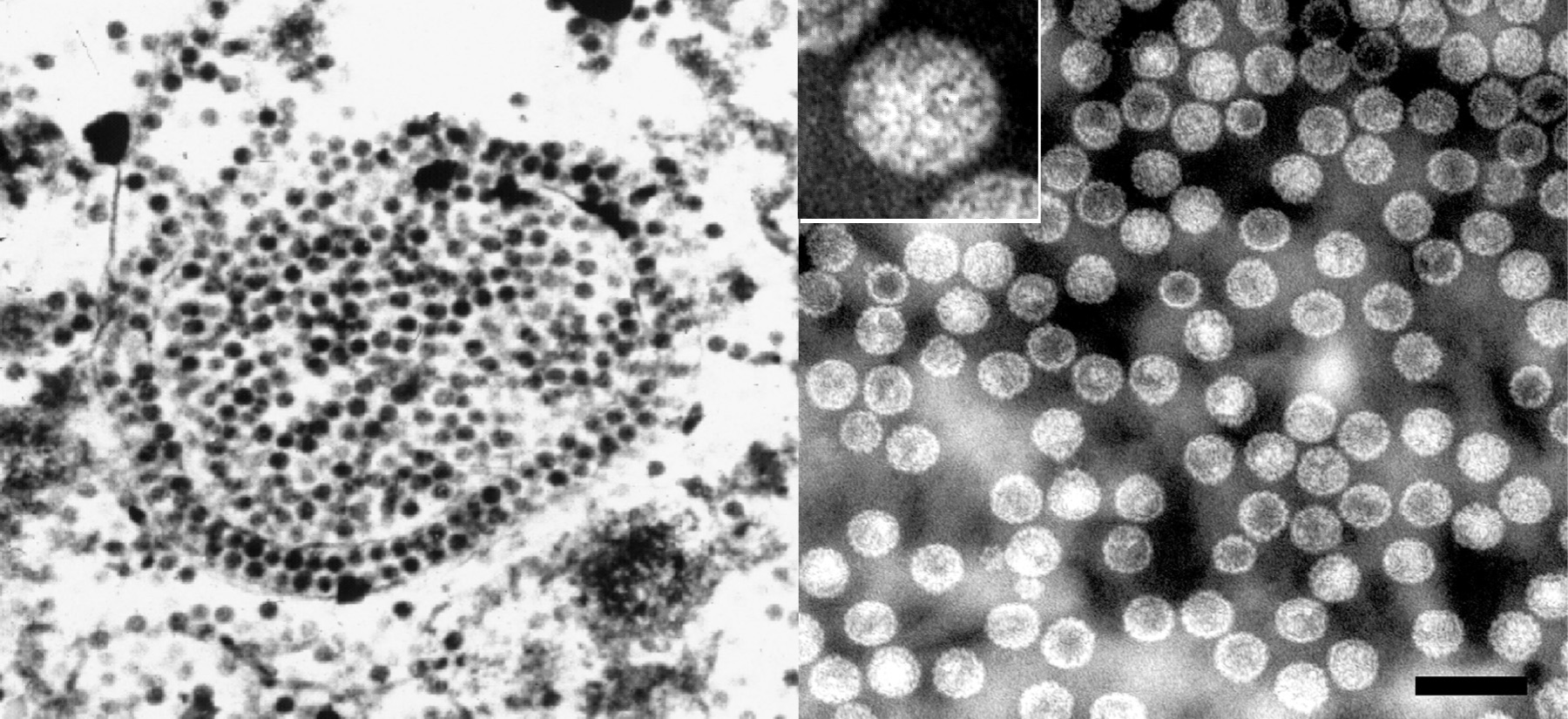
Figure 3 Diagram of representative polyomavirus genomes and encoded proteins, SV40 (left) and MPyV (right). The grey circles represent the viral dsDNAs. The origin of DNA replication (ORI) is indicated. Arrows indicate protein-coding regions and their direction of transcription. Introns are denoted by solid lines. Note that the non-coding region in the vicinity of ORI also includes the regulatory region. Alternative splicing is a common characteristic of polyomavirus coding regions. The multiple large T gene products all share identical amino termini.
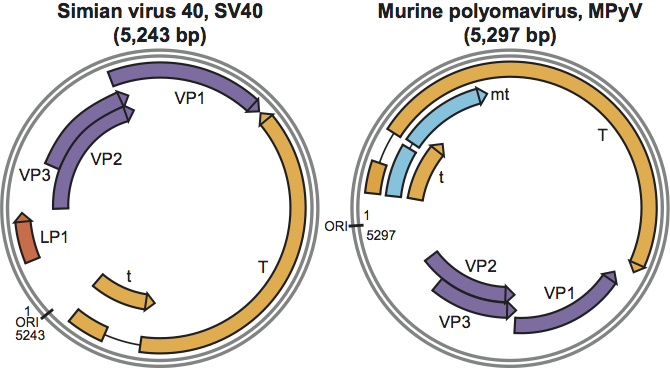
Figure 4 The Compass: A schematic diagram of the relationships between JC polyomavirus (JCPyV) regulatory region sequences published worldwide. JCPyV variant regulatory regions are grouped into quadrants (I-S, I-R, II-S and II-R) with ace sequence-units lightly shaded. Upper quadrant variant types (I) have no additional sequence integrated into the ace units (no inserts). Lower quadrant variant types (II) have dark integrated sequence sections (inserts), b (23 bp) and d (66 bp). Both types I and II are divided into singular (S) and repeat (R) forms by the left and right quadrants, respectively. Unshaded boxes are TATA boxes. Dots represent sites of possible deletions. Unshaded diamonds contain the nucleotide that occupies the 49th position in sequence section c (nt 85 of I-S, or 108 of II-S), which is adenine (A) in type I variants, but predominantly guanine (G) in type II variants. Right quadrants (R-forms) have dark dashes where sequence is deleted and where additional repeats may occur. The * in the lower right quadrant (II-R) identifies one reported sequence that retains the second TATA box (Ciappi et al., 1999). JCPyV tropism common to all variant regulatory region forms is contained in the dark central circle. Specific JCPyV tropisms are contained in the dark corner triangles. Cells from tonsil are either (L) lymphocytes, or (S) stromal cells (Monaco et al., 1998). Cells in bone marrow that contain JCPyV have been identified as B-lymphocytes
(Houff et al., 1988).
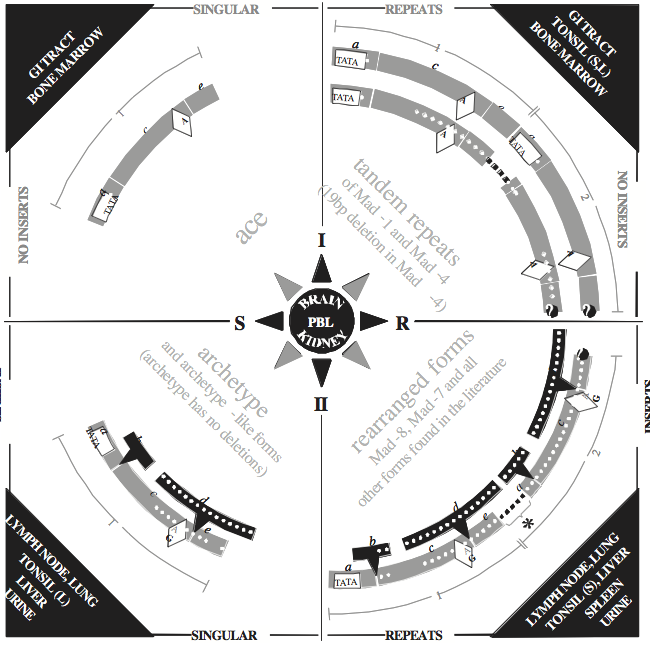
Figure 5 Phylogentic relationships of polyomaviruses. The figure shows a concatenated data set of maximum likelihood (ML) trees that were obtained for each of VP1, VP2 and T antigen. Branch lengths are proportional to genetic divergence. The scale bars indicate nuclear substitutions per site.
(After Krumbholz, A. et al. (2009). Infect. Genet. Evol., 9, 784799.)