Family: Polydnaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions have a complex construction, consisting of a nucleocapsid and a single or double layer envelope. Virions consist of one or more enveloped nucleocapsids. Polydnaviruses are divided into two genera, Bracovirus and Ichnovirus, which share few morphological features (Figure 1). Morphological traits for each genus are described below.
Physicochemical and physical properties
None reported.
Nucleic acid
Encapsidated genomes of polydnaviruses consist of multiple dsDNAs of variable size (Figure 2). Genome segments are non-equimolar in abundance. The encapsidated form of polydnavirus genomes is segmented; segments consist of circular supercoiled double stranded DNA. Virions may also contain homologous DNA sequences that are shared among two or more DNA genome segments. Aggregate, non-redundant, genome sizes range from approximately 190 to more than 500 kbp.
Proteins
Virions are structurally complex and contain at least 20–30 polypeptides, with sizes ranging from 10 to 200 kDa.
Lipids
Lipids are present, but uncharacterized.
Carbohydrates
Carbohydrates are present, but uncharacterized.
Genome organization and replication
Unique among dsDNA viruses, polydnaviruses are specifically associated with parasitoid wasps in the insect order Hymenoptera. Each polydnavirus carried by a given wasp species is genetically unique and exists in two forms. Polydnaviruses persist and are transmitted from adult wasp to offspring as proviruses that are stably integrated into the genome of the wasp. Replication, which results in production of the encapsidated form of the virus, is restricted to specialized calyx cells in the ovaries of female wasps. Replication in calyx cells is nuclear and begins during wasp pupal-adult development. Virus morphogenesis occurs in calyx cells of all female wasps. Viral DNA replication involves amplification of genes plus proviral DNAs corresponding to the viral DNAs packaged into particles. Proviral DNAs are excised via site-specific recombination events and packaged into virus particles, whereas amplified genes required for particle formation are not. Bracovirus particles are released by lysis of calyx cells, while ichnovirus particles bud from calyx cells. Both ichno- and bracoviruses accumulate to high density in the lumen of the oviducts, and wasps inject a quantity of these particles into host insects at oviposition (Figure 3). Shared features of the encapsidated form of polydnavirus genomes include low coding densities and strong A+T biases. A majority of predicted genes in the encapsidated genome consist of related variants, which form multimember gene families. Several genes also contain introns. Transcriptional activity of polydnaviruses is host-specific with some genes expressed only in the wasp, other genes expressed only in the parasitized host of the wasp, and a few genes expressed in both the wasp and parasitized hosts. Bracoviruses and ichnoviruses share few genes or gene families with one another due to their distinct evolutionary origins (see below). Shared genomic and biological features, therefore, reflect convergent evolution. It is also possible that rare genetic exchanges have occurred between polydnaviruses carried by different wasps that parasitize the same host species.
Antigenic properties
Members of a number of different Ichnovirus species share cross-reacting antigenic determinants; in some cases, viral nucleocapsids share at least one major conserved epitope. Campoletis sonorensis ichnovirus (CsIV) and C. sonorensis venom proteins display some common epitopes. Although less understood, bracoviruses also share some antigenic determinants with one another.
Biological properties
All polydnavirus-carrying wasps reside in two families (Braconidae and Ichneumonidae) of the Hymenoptera. All wasp species lay (oviposit) their eggs inside the body of hosts, which are primarily other insects in the order Lepidoptera (Figure 3). Wasp offspring develop by feeding on host tissues. Polydnaviruses are transmitted only vertically from wasp to offspring as proviruses (Figure 3). Reciprocally, wasps infect hosts with only the encapsidated form of polydnaviruses. After parasitism, virus particles infect host tissues and multiple viral genes are thereafter expressed (Figure 3). However, polydnaviruses do not replicate in the wasp’s host due to the absence of genes required for particle formation (see above). Virus-specific gene products cause significant changes in the physiology of the wasp’s host, which are required for successful development of offspring. Thus, a mutualism exists because transmission as a provirus depends upon survival of the wasp, and survival of the wasp depends upon infection of its host by the encapsidated, non-replicating form of the virus.
Genus Bracovirus
Type species Cotesia melanoscela bracovirus
Distinguishing features
Bracoviruses are associated with an estimated 18,000 species of wasps in five subfamilies of the Braconidae (Microgastrinae, Cardiochilinae, Miracinae, Khoikholinae and Cheloninae) which together form a monophyletic assemblage called the microgastroid complex. Bracovirus nucleocapsids are cylindrical and surrounded by a single unit membrane envelope. Bracovirus virions contain either single or multiple nucleocapsids in a species-dependent manner.
Virion properties
Morphology
Virions consist of enveloped cylindrical electron-dense nucleocapsids of uniform diameter (34–40 nm) but of variable length (8–150 nm length) and may contain one or more nucleocapsids within a single envelope assembled de novo in the nuclei of calyx cells. Bracovirus nucleocapsids also possess long unipolar tail-like appendages (Figure 4).
Nucleic acid
The encapsidated genomes of bracoviruses consist of multiple, circular dsDNAs ranging in size from approximately 2.0 to more than 30 kbp. Aggregate, non-redundant genome sizes range from 189 kbp for Microplitis demolitor bracovirus (MdBV) to more than 600 kbp for Cotesia congregata bracovirus (CcBV). Genomic segments vary from as few as six for Toxoneuron nigriceps bracovirus (TnBV) to 29 DNAs for Glyptapanteles indiensis bracovirus (GiBV). The corresponding proviral (linear) form of bracovirus genomes reside at multiple macroloci located in specialized regions of the wasp genome. Some of the genes required for replication and the formation of particles (i.e. viral machinery) also cluster in a unique region(s) in the wasp genome.
Genome organization and replication
The proviral genome segments cluster in tandem arrays in specialized regions of the wasp genome (Figure 5). Proviral DNA is amplified in calyx cells of female wasp ovaries at the onset of replication in the pupal stage. Genomic segments are excised from the amplified cluster at repetitive sequences for packaging into virions. In the case of Glyptapanteles indiensis bracovirus (GiBV), 20 of the 29 segments comprising the encapsidated form of the genome form one macrolocus, while five other proviral loci contain either a single or two proviral segments in tandem array. Small, intersegmental regions (<1kbp), lacking any predicted open reading frames, separate the tandemly arrayed proviral segments. Approximately half of the genes required for particle formation, related to structural genes of nudiviruses, are also clustered in the wasp genome, albeit not in close proximity to proviral loci. At the onset of replication, sequences corresponding to proviral loci are amplified in calyx cells, while genes involved in particle formation are expressed at high levels. Amplified DNAs are then excised and packaged into virus particles, whereas amplified genes required for particle formation are not. Genomic segments are individually packaged into particles and are non-equimolar in abundance. Most genomic segments packaged into particles encode one or more genes but coding densities are overall low. A majority of genes belong to multimember gene families. Gene family members may be clustered on a single viral genomic segment or distributed on multiple segments. Bracoviruses associated with closely related wasps appear to encode similar genes and gene families, whereas the encapsidated genomes of bracoviruses from distantly related wasps share few or no genes. Core genes involved in particle formation in contrast appear conserved among bracoviruses.
Antigenic properties
Antigenic relationships among the bracoviruses have not been investigated in detail although cross-reacting epitopes have been detected.
Biological properties
Braconid wasps that carry bracoviruses form a monophyletic lineage. All known bracovirus-carrying wasps inject a quantity of virus particles into hosts during oviposition. For most species, virus particles preferentially infect host immune cells (hemocytes) plus selected other tissues including the fat body. Expression of viral gene products in the absence of replication leads to significant changes in host physiology essential for successful parasitism. Bracoviruses from several wasp species prevent the immune system of the host from killing the wasp’s offspring. Several immunosuppressive genes encoded by bracovirus have been identified. Bracovirus gene products also alter host development and metabolism.
Species demarcation criteria in the genus
To demonstrate experimentally that a virus is a bracovirus, the following criteria should be met:
- Virions were isolated from the reproductive tract (calyx region) of adult, female braconid wasps
- Virions have bracovirus morphology, including cylindrical nucleocapsids of variable length, with a diameter of about 30 nm surrounded by a single unit membrane, single or multiple nucleocapsids may be present in the virion
- Nucleic acid isolated from virions is circular dsDNA from multiple molecules (i.e. the DNA genome is segmented)
- Reference specimens of the wasp host are identified by a qualified systematics specialist and deposited in an accessible insect collection.
Criteria that are thought to be of systematic significance:
- Parasitoid wasp species (proviral and replicative host)
- Host(s) of the wasp and its associated bracovirus (the non-replicative host)
- DNA restriction map profiles and/or sequence.
List of species in the genus Bracovirus
| Apanteles crassicornis bracovirus |
|
|
| Apanteles crassicornis bracovirus |
| (AcBV) |
| Apanteles fumiferanae bracovirus |
|
|
| Apanteles fumiferanae bracovirus |
| (AfBV) |
| Ascogaster argentifrons bracovirus |
|
|
| Ascogaster argentifrons bracovirus |
| (AaBV) |
| Ascogaster quadridentata bracovirus |
|
|
| Ascogaster quadridentata bracovirus |
| (AqBV) |
| Cardiochiles nigriceps bracovirus |
|
|
| (Toxoneuron nigriceps bracovirus) |
|
|
| Toxoneuron nigriceps bracovirus | [Y19010, AJ440973] | (TnBV) |
| Chelonus altitudinis bracovirus |
|
|
| Chelonus altitudinis bracovirus |
| (CalBV) |
| Chelonus blackburni bracovirus |
|
|
| Chelonus blackburni bracovirus |
| (CbBV) |
| Chelonus inanitus bracovirus |
|
|
| Chelonus inanitus bracovirus | [Z31378, Z58828, AM261417, AM850131, AJ319653, AJ278673, AJ627175] | (CiBV) |
| Chelonus insularis bracovirus |
|
|
| Chelonus insularis bracovirus |
| (CinsBV) |
| Chelonus nr. curvimaculatus bracovirus |
|
|
| Chelonus nr. curvimaculatus bracovirus |
| (CcvBV) |
| Chelonus texanus bracovirus |
|
|
| Chelonus texanus bracovirus |
| (CtBV) |
| Cotesia congregata bracovirus |
|
|
| Cotesia congregata bracovirus | [AF049877, AJ632304, EU493286, D29821, AJ640087, AM180416, AJ583542, AF049876, AF006205] | (CcBV) |
| Cotesia flavipes bracovirus |
|
|
| Cotesia flavipes bracovirus | [EU493300, EU493297] | (CfBV) |
| Cotesia glomerata bracovirus |
|
|
| Cotesia glomerata bracovirus | [FJ713017, AY481559, AY486078, AY466396, EU493327, DQ844603, DQ839630] | (CgBV) |
| Cotesia hyphantriae bracovirus |
|
|
| Cotesia hyphantriae bracovirus |
| (ChBV) |
| Cotesia kariyai bracovirus |
|
|
| Cotesia kariyai bracovirus | [AB099714, AB086812, AB074136] | (CkBV) |
| Cotesia marginiventris bracovirus |
|
|
| Cotesia marginiventris bracovirus |
| (CmaBV) |
| Cotesia melanoscela bracovirus |
|
|
| Cotesia melanoscela bracovirus | [EU493303] | (CmeBV) |
| Cotesia rubecula bracovirus |
|
|
| Cotesia rubecula bracovirus | [U55279, AY631272, AY234855, AF359344, EU493316] | (CrBV) |
| Cotesia schaeferi bracovirus |
|
|
| Cotesia schaeferi bracovirus |
| (CsBV) |
| Diolcogaster facetosa bracovirus |
|
|
| Diolcogaster facetosa bracovirus |
| (DfBV) |
| Glyptapanteles flavicoxis bracovirus |
|
|
| Glyptapanteles flavicoxis bracovirus |
| (GflBV) |
| Glyptapanteles indiensis bracovirus |
|
|
| Glyptapanteles indiensis bracovirus | [EF051505, AY871265, AY162267, AF414845, EU001243, AF198385] | (GiBV) |
| Glyptapanteles liparidis bracovirus |
|
|
| Glyptapanteles liparidis bracovirus |
| (GlBV) |
| Hypomicrogaster canadensis bracovirus |
|
|
| Hypomicrogaster canadensis bracovirus |
| (HcBV) |
| Hypomicrogaster ectdytolophae bracovirus |
|
|
| Hypomicrogaster ectdytolophae bracovirus |
| (HecBV) |
| Microplitis croceipes bracovirus |
|
|
| Microplitis croceipes bracovirus |
| (McBV) |
| Microplitis demolitor bracovirus |
|
|
| Microplitis demolitor bracovirus | [AF267174, U76033, AY842013, AY848690, AY875680, AY887894, DQ000240] | (MdBV) |
| Phanerotoma flavitestacea bracovirus |
|
|
| Phanerotoma flavitestacea bracovirus |
| (PfBV) |
| Pholetesor ornigis bracovirus |
|
|
| Pholetesor ornigis bracovirus |
| (PoBV) |
| Protapanteles paleacritae bracovirus |
|
|
| Protapanteles paleacritae bracovirus |
| (PpBV) |
| Tranosema rostrale bracovirus |
|
|
| Tranosema rostrale bracovirus |
| (TrBV) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Bracovirus but have not been approved as species
| Cotesia chilonis bracovirus | [EU493292-6] | (CchBV) |
| Cotesia plutellae bracovirus | [AY651828-30, AY461733, DQ858218-9, EU523381, DQ299488, DI146230, DI145340, DI143592, DI112181, DI111256, DI114377, DI112939, DI110650, EF067331-2, EF067319-30, DQ075354-60] | (CpBV) |
| Cotesia ruficrus bracovirus | [AB099713] | (CrfBV) |
| Cotesia sesamiae bracovirus | [EF710626-35, EF710636-43] | (CseBV) |
| Cotesia vestalis bracovirus | [EU127911, FJ176776-8, EU493308-10, EU081840, EU095951, EF467277-8] | (CvBV) |
| Microplitis bicoloratus bracovirus | [DQ286649] | (MbBV) |
| Toxoneuron nigriceps bracovirus | [Y19010, AJ440973] | (MdBV) |
Genus Ichnovirus
Type species Campoletis sonorensis ichnovirus
Distinguishing features
Ichnoviruses are associated with an estimated 13,000 species of wasps in two subfamilies of the Ichneumonidae (Campopleginae and Banchinae). Ichnovirus nucleocapsids are fusiform or quasi-cylindrical, often with a short tail-like appendage, and enveloped by two unit membranes. Virions associated with campoplegine ichneumonids contain a single nucleocapsid, whereas virions associated with banchine ichneumonids envelope multiple nucleocapsids.
Virion properties
Morphology
Ichnovirus virions consist of nucleocapsids of uniform size (approximately 85×330 nm), having the form of a prolate ellipsoid, surrounded by two unit membrane envelopes (Figure 6). The inner envelope appears to be assembled de novo in the nuclei of calyx cells, while the outer envelope is acquired by budding through the plasma membrane of calyx cells.
Nucleic acid
The encapsidated genomes of ichnoviruses consist of multiple, circular dsDNAs ranging in size from approximately 2.0 to more than 25 kbp. Ichnoviruses carried by campoplegine ichneumonids like Campoletis sonorensis ichnovirus (CsIV) and Hyposoter fugitivus ichnovirus consist of 20–25 genomic segments with an aggregate size of approximately 250 kbp. In contrast, ichnoviruses associated with banchine ichneumonids, like Glypta fumiferanae virus (GfIV) has an aggregate size of approximately 290 kbp divided into more than 100 segments that range from 1.5 to 5.0 kbp.
Genome organization and replication
Ichnoviruses replicate from proviral DNA. Proviral DNA is amplified in calyx cells of female wasp ovaries at the onset of replication in the pupal stage. Genomic segments are excised from the amplified cluster at repetitive sequences for packaging into virions. Genes required for particle formation are clustered in the wasp genome, forming multiple ichnovirus structural protein-encoding regions. In the wasp Hyposoter didymator, two of these structural protein-encoding regions are located in proximity to proviral loci. At the onset of replication, structural protein-encoding regions and proviral loci amplify in calyx cells. Proviral DNAs are then excised and packaged into virus particles, but structural protein-encoding regions are not. Encapsidated ichnovirus genomes are non-equimolar with two recognized viral segment types, nested and unique (Figure 7). Nested segments are often hypermolar and produce from 2 to 5 partially redundant segments from a single proviral locus. Unique segments excise to produce a single segment that is encapsidated. There is evidence that nested segments are preferentially associated with some gene families. Most genomic segments encode one or more genes but coding densities are overall low. A majority of genes belong to multimember gene families. Gene family members may be clustered on a single viral segment or distributed on multiple segments. The encapsidated genomes of ichnoviruses from campoplegine ichneumonids share six gene families, but only one of these gene families is shared with ichnoviruses from banchine ichneumonids. Several genes involved in particle formation are conserved among ichnoviruses, but are unrelated to structural-protein encoding genes of bracoviruses.
Antigenic properties
Cross-reacting antigenic determinants are shared by a number of different ichnovirus isolates; in some cases, viral nucleocapsids share at least one major conserved epitope. CsIV and C. sonorensis venom protein display common epitopes.
Biological properties
Ichnovirus-carrying wasps inject a quantity of virus particles into host animals during oviposition; virus-specific expression leads to significant changes in host physiology, some of which are responsible for successful parasitism. Ichnoviruses inhibit the immune responses and alter development of infected (parasitized) hosts. Infection impacts translation of some host mRNAs and this impacts the ability of the host to mount effective immune responses (e.g. melanization).
Species demarcation criteria in the genus
To demonstrate experimentally that a virus is an ichnovirus, the following criteria should be met:
- Virions were isolated from the reproductive tract (calyx region) of adult, female ichneumonid wasps
- Virions have ichnovirus morphology including fusiform nucleocapsids surrounded by a double unit membrane, single or multiple nucleocapsids may be present per virion
- Nucleic acid isolated from virions is circular dsDNA from multiple molecules (i.e. the DNA genome is segmented)
- Reference specimens of the wasp host identified by a qualified systematics specialist and deposited in an accessible insect collection.
Criteria that are thought to be of systematic significance:
- Parasitoid wasp (proviral and replicative host)
- Host(s) of the wasp and its associated ichnovirus (the non-replicative host)
- DNA restriction map profiles and/or sequence.
List of species in the genus Ichnovirus
| Campoletis aprilis ichnovirus |
|
|
| Campoletis aprilis ichnovirus | [FJ463032, DQ845287-8, AB100268] | (CaIV) |
| Campoletis falvincta ichnovirus |
|
|
| Campoletis flavicincta ichnovirus |
| (CfIV) |
| Campoletis sonorensis ichnovirus |
|
|
| Campoletis sonorensis ichnovirus | [U41655, S47226, AY029394, AY029400, AF361487, AF411011, AF362507, AF361869, AF004367, AY573925, AY197485, AF236017, AF004366, AF004378, L08243, M17405, M23437, M80621-3, AY953130, AH006861, AF004557, M17004, M16999, M17404] | (CsIV) |
| Casinaria arjuna ichnovirus |
|
|
| Casinaria arjuna ichnovirus |
| (CarIV) |
| Casinaria forcipata ichnovirus |
|
|
| Casinaria forcipata ichnovirus |
| (CfoIV |
| Casinaria infesta ichnovirus |
|
|
| Casinaria infesta ichnovirus |
| (CiIV |
| Diadegma acronyctae ichnovirus |
|
|
| Diadegma acronyctae ichnovirus |
| (DaIV) |
| Diadegma interruptum ichnovirus |
|
|
| Diadegma interruptum ichnovirus |
| (DiIV) |
| Diadegma terebrans ichnovirus |
|
|
| Diadegma terebrans ichnovirus |
| (DtIV) |
| Enytus montanus ichnovirus |
|
|
| Enytus montanus ichnovirus |
| (EmIV) |
| Eriborus terebrans ichnovirus |
|
|
| Eriborus terebrans ichnovirus |
| (EtIV) |
| Glypta fumiferanae ichnovirus |
|
|
| Glypta fumiferanae ichnovirus | [AB295392, AB289903-99, AB290000-7] | (GfIV) |
| Hyposoter annulipes ichnovirus |
|
|
| Hyposoter annulipes ichnovirus |
| (HaIV) |
| Hyposoter exiguae ichnovirus |
|
|
| Hyposoter exiguae ichnovirus |
| (HeIV) |
| Hyposoter fugitivus ichnovirus |
|
|
| Hyposoter fugitivus ichnovirus | [AY597814, AY577428-9, AY570798-9, AY563518-9, AY556383-4, AY547319, AB291200-9, AB291165-99, AY935249] | (HfIV) |
| Hyposoter lymantriae ichnovirus |
|
|
| Hyposoter lymantriae ichnovirus |
| (HlIV) |
| Hyposoter pilosulus ichnovirus |
|
|
| Hyposoter pilosulus ichnovirus |
| (HpIV) |
| Hyposoter rivalis ichnovirus |
|
|
| Hyposoter rivalis ichnovirus |
| (HrIV) |
| Olesicampe benefactor ichnovirus |
|
|
| Olesicampe benefactor ichnovirus |
| (ObIV) |
| Olesicampe geniculatae ichnovirus |
|
|
| Olesicampe geniculatae ichnovirus |
| (OgIV) |
| Synetaeris tenuifemur ichnovirus |
|
|
| Synetaeris tenuifemur ichnovirus |
| (StIV) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Ichnovirus but have not been approved as species
| Campoletis chlorideae ichnovirus | [FJ463032, DQ845287-8, AB100268] | (CaIV) |
| Campoletis sp. ichnovirus | [AY033945] | (CspIV) |
| Casinaria sp. ichnovirus |
| (CaspIV) |
| Dusona sp. ichnovirus |
| (DspIV) |
| Glypta sp. ichnovirus |
| (GspIV) |
| Hyposoter didymator ichnovirus | [AF132024, DQ295918-20, AF241775, AY519505-6, AY518195-9, AY499565-9, AF479654, AF464931, AF364055-7, AF191723, AF132023, AF131648, AF237946, AF156933, AF464930, AY501381-3, AY486462-4] | (HdIV) |
| Lissonota sp. ichnovirus |
| (LspIV) |
| Tranosema rostrale ichnovirus | [DQ790660, DQ790662-3, AF052836-7, AF529168, AF527780, AB291213-5, AB291138-64, AF421353, AY940454] | (TrIV) |
Phylogenetic relationships within the family
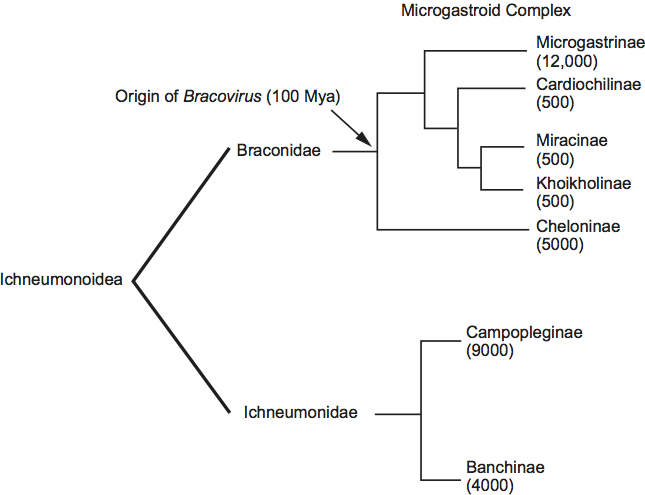
Phylogenetic data indicate all members of the Polydnaviridae are associated with two families of wasps (Braconidae and Ichneumonidae) in the superfamily Ichneumonoidea (Figure 8). However, the members of the genera Bracovirus and Ichnovirus are unrelated by sequence and serological analyses. All bracoviruses are associated with wasps in specific subfamilies of the family Braconidae. Together, these subfamilies form a monophyletic lineage referred to as the microgastroid complex (Figure 8). Phylogenetic data and fossil calibrations indicate that the microgastroid complex arose approximately 100 million years ago (Mya) and that the bracovirus–braconid wasp association arose from an interaction established between a single ancestral nudivirus and the common ancestor of the microgastroid complex. This interaction then diversified into the thousands of bracovirus-carrying braconid wasp species that exist today. All ichnoviruses are associated with wasps in two subfamilies of the family Ichneumonidae (Figure 8). Phylogenetic analysis indicates that the two families of ichnovirus carrying ichneumonid wasps are separated by groups that do not carry ichnoviruses, which suggest an independent acquisition of viruses in the two groups or a loss of PDVs in some groups. Taken together, dissimilar morphologies and genomes indicate that bracoviruses and ichnoviruses do not share a common ancestor. Instead, ichnoviruses and bracoviruses are currently classified in the family Polydnaviridae, because of: (1) similarities in life cycle and (2) both produce replicatively defective encapsidated genomes that are comprised of multiple circular, dsDNAs. These similarities, however, reflect convergent evolution.
Similarity with other taxa
Sequence analysis of genes required for particle formation indicate that bracoviruses evolved from an ancestral nudivirus. Sequence analysis of genes required for particle formation suggest ichnoviruses also arose from a viral ancestor. Evidence strongly indicates this ancestor was not a nudivirus but a lack of homology with any known taxon of viruses suggests ichnoviruses originated from a virus ancestor now extinct or from a currently undiscovered taxon.
Derivation of names
Braco: from Braconidae, a family of wasps.
Ichno: from Ichneumonidae, a family of wasps.
Polydna: from Greek poly, “several”, and dna, deoxyribonucleic acid.
Further reading
Bezier, A., Annaheim, M., Herbiniere, J., Wetterwald, G., Gyapay, S., Bernard-Samain, S., Wincker, P., Rodidi, I., Heller, M., Belghaazi, M., Pfister-Wilhem, R., Periquet, G., Dupuy, C., Huguet, E., Volkoff, A.N., Lanzrein, B. and Drezen, J.-M. (2009). Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science, 323, 926-930.
Desjardins, C.A., Gundersen-Rindal, D.E., Hostetler, J.B., Tallon, L.J., Fadrosh, D.W., Fuester, R.W., Pedroni, M.J., Haas, B.J., Scatz, M.C., Jones, K.M., Crabtree, J., Forgerger, H. and Nene, V. (2008). Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps. Genome Biol., 9, R183.
Dupuy, C., Huguet, E., and Drezen, J.-M. (2006). Unfolding the evolutionary history of polydnaviruses. Virus Res., 117, 81-89.
Espagne, E., Dupuy, C., Huguet, E., Cattolico, L., Provost, B., Martins, N., Poirie, M., Periquet, G. and Drezen, J.-M. (2004). Genome sequence of a polydnavirus: insights into symbiotic virus evolution. Science, 306, 286-289.
Lapointe, R., K. Tanaka, W. E. Barney, W.E., Whitfield, J.B., Banks, J.C., Beliveau, C., Stoltz, D., Webb, B.A. and Cusson, M. (2007). Genomic and morphological features of a banchine polydnavirus: comparison with bracoviruses and ichnoviruses. J. Virol., 81, 6491-6501.
Strand, M. R. (2009). The interactions between polydnavirus-carrying parasitoids and their lepidopteran hosts. In M.R. Goldsmith and F. Marec (Eds.), Molecular Biology and Genetics of the Lepidoptera, Boca Raton, FL, CRC Press, pp. 321-336.
Volkoff, A.N., Jouan, V., Urbach, S., Samain, S., Bergoin, M., Wincker, P., Demettre, E., Cousserans, F., Provost, B., Coulibaly, F., Legeai, F., Beliveau, Ca., Cusson, M., Gyapay, G. and Drzen, J.-M. (2010). Analysis of virion structural components reveals vestiges of the ancestral ichnovirus genome. PLOS Pathog. 6, e1000923.
Webb, B.A. and Strand, M.R. (2005). The biology and genomics of polydnaviruses. In: L.I. Gilbert, K. Iatrou and S.S. Gill (Eds.), Comprehensive Molecular Insect Science, Vol. 6. San Diego, CA: Elsevier, pp. 323-360.
Webb, B.A., Strand, M.R., Deborde, S.E., Beck, M., Hilgarth, R.S., Kadash, K., Kroemer, J.A., Lindstrom, K.G., Rattanadechakul, W., Shelby, K.S., Thoetkiattikul, L., Turnbull, M.W., Barney, W.E. and Witherell, R.A. (2006). Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology, 347, 160-174.
Whitfield, J.B. (2002). Estimating the age of the polydnavirus/braconid wasp symbiosis. Proc. Natl Acad. Sci. USA, 99, 7508-7513.
Contributed by
Strand, M.R. and Drezen, J.-M.
Figures
Figure 1 Cotesia melanoscela bracovirus (left) and Campoletis sonorensis ichnovirus virions (right). The bars represent 200 nm.
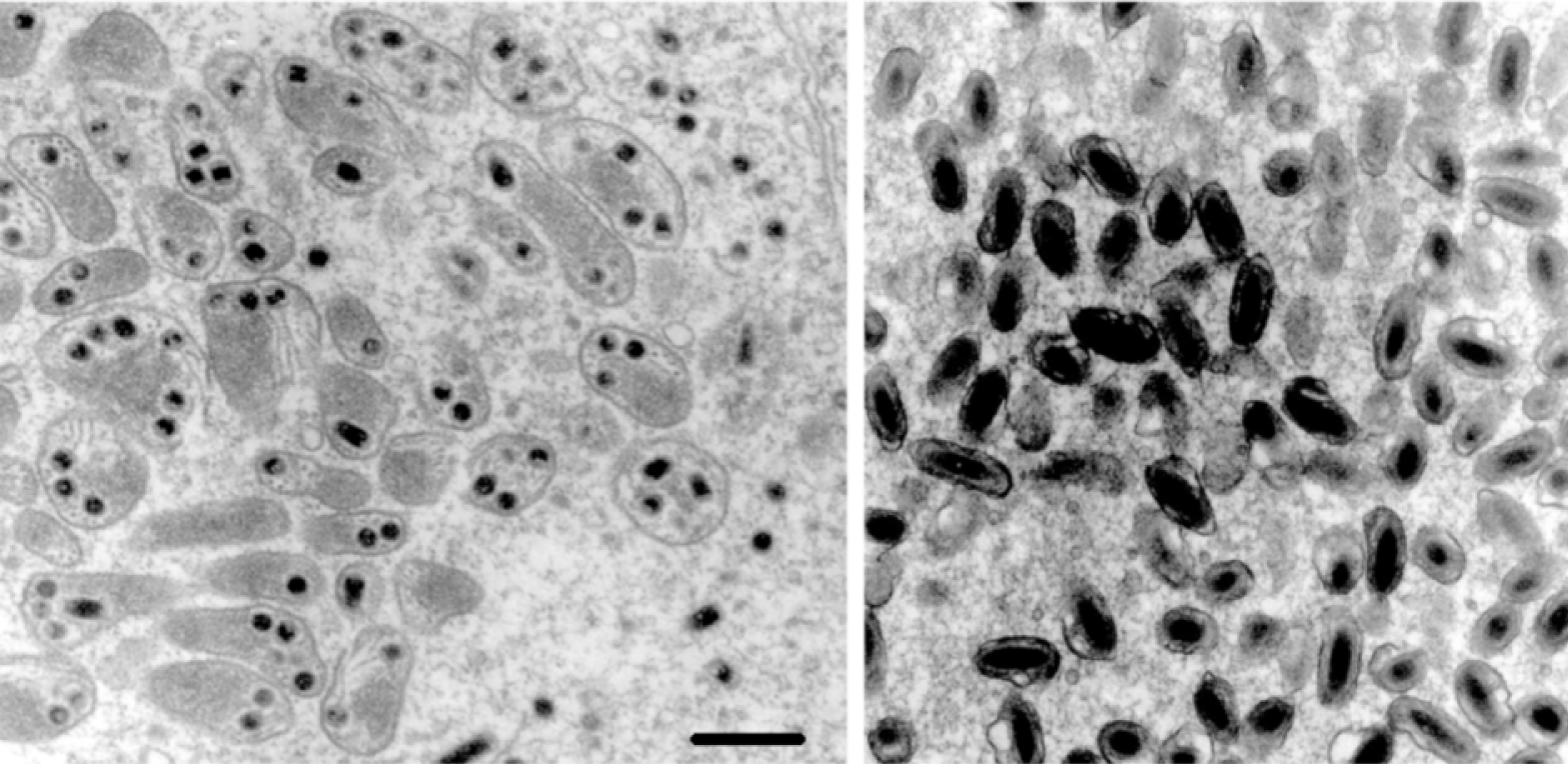
Figure 2 Encapsidated DNA genomes from a member of the genus Ichnovirus (Campoletis sonorensis ichnovirus, CsIV); left panel, two lanes with two different amounts of DNA); and a Bracovirus (Cotesia marginiventris bracovirus, CmaBV; right panel, two lanes with two different amounts of DNA). Genomes were electrophoresed on 1% agarose gels and visualized with ethidium bromide. The different amounts of DNA illustrate the non-equimolarity of individual genomic segments.
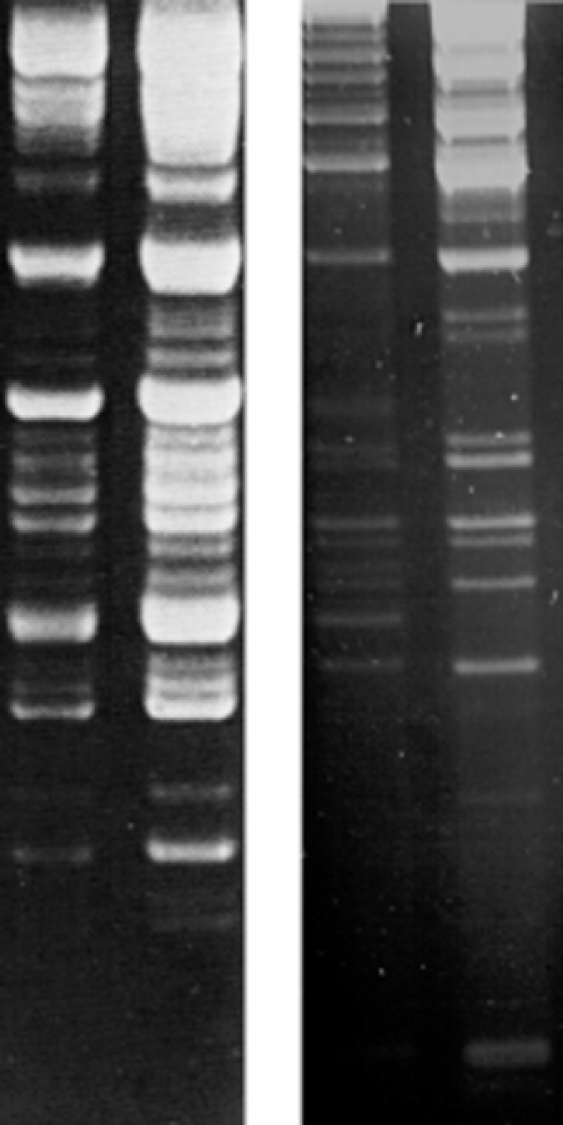
Figure 3 Polydnavirus replication and transmission cycles. The replication and transmission of polydnaviruses and the life cycle of an endoparasitic wasp are illustrated. Viral DNA is transmitted as proviral DNA in wasp cells (thin arrows) and as circular episomal DNAs within virions (thick arrows). In replicative wasp cells and in infected lepidopteran cells, viral DNA is present in an unpackaged closed circular form. In non-replicative cells (i.e. all wasp cells except female pupal/adult calyx cells) the virus does not replicate and exists predominantly in the proviral form. Polydnaviruses are vertically transmitted in only the proviral DNA form.
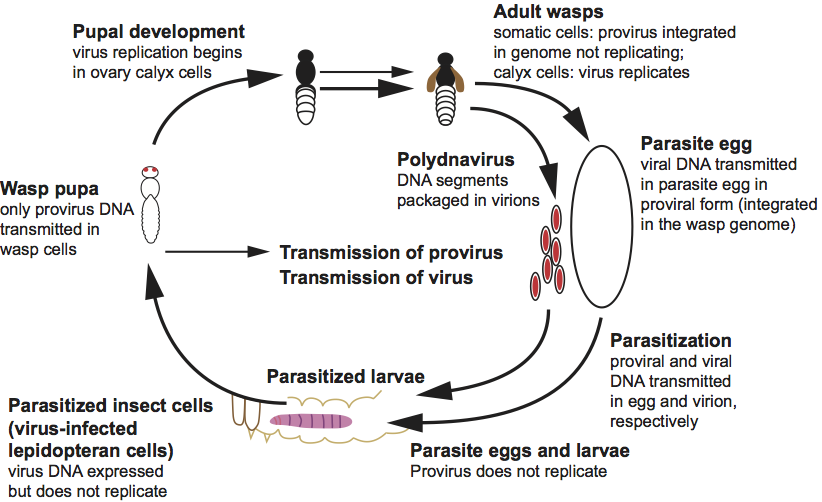
Figure 4 (Left) Sectional diagram and (right) negative contrast electron micrograph of particles of Protapanteles paleacritae bracovirus (PpBV). The bar represents 200 nm.
(Courtesy of D. Stoltz.)
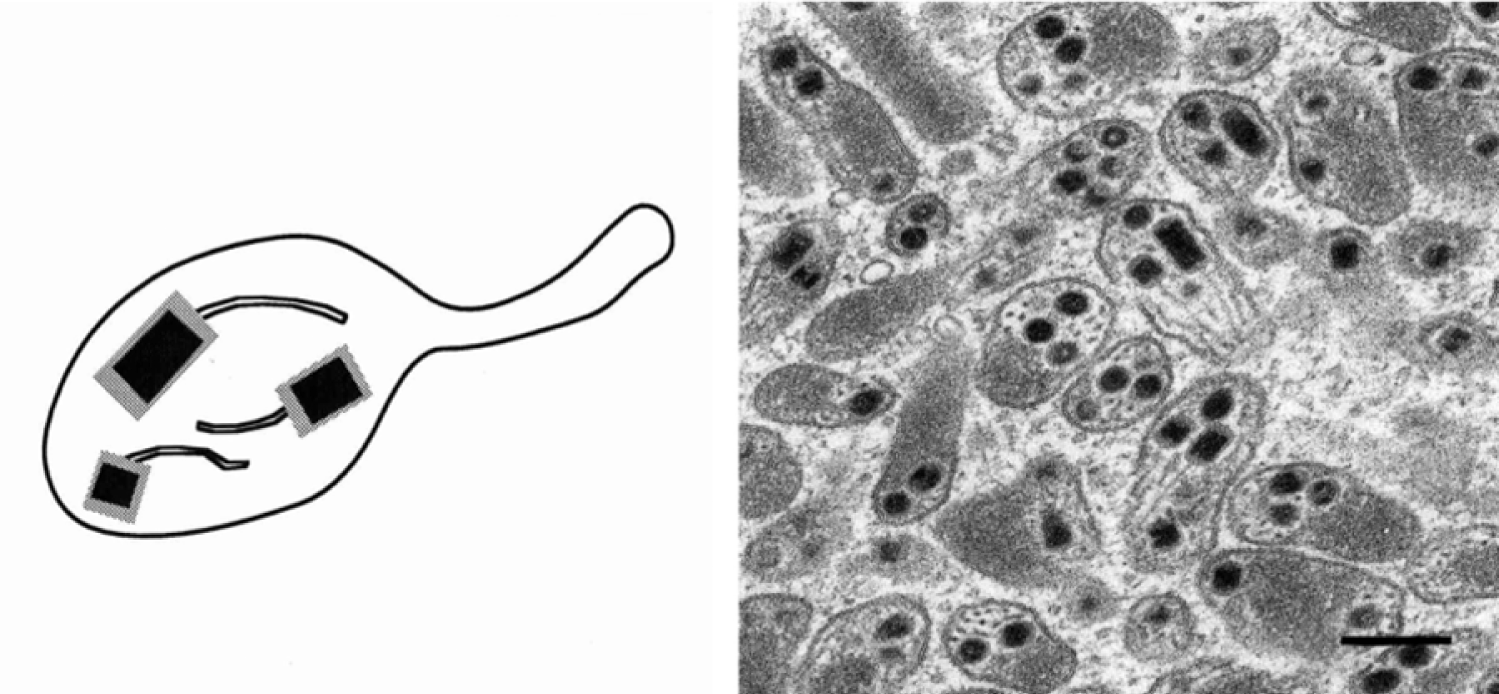
Figure 5 Bracovirus genome organization and replication. The Microplitis demolitor bracovirus (MdBV) genome is shown on left with selected viral segments identified. The right panel shows a representative bracovirus segment, MdBV segment K, in the proviral (top) and episomal form (bottom). Proviral segments may be found in tandem arrays with adjacent viral segments (AVS) flanking the integration site of proviral segments. MdBV segments encode genes but are predominantly non-coding sequence.
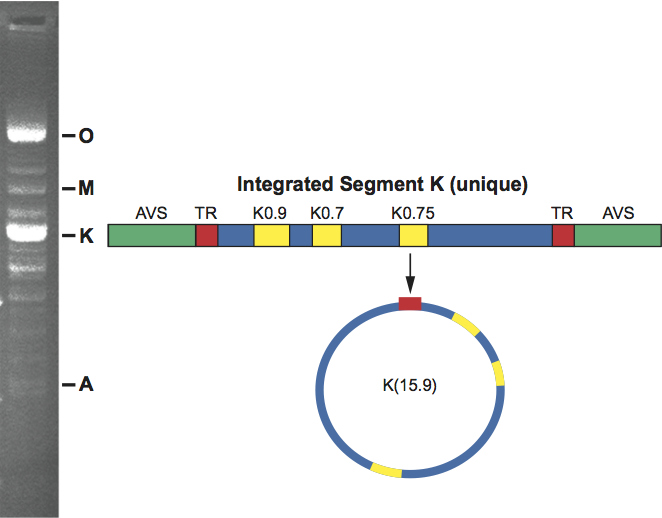
Figure 6 (Left) Sectional diagram and (right) negative contrast electron micrograph of particles of Hyposoter exiguae ichnovirus (HeIV). The bar represents 200 nm.
(Courtesy of D. Stoltz.)
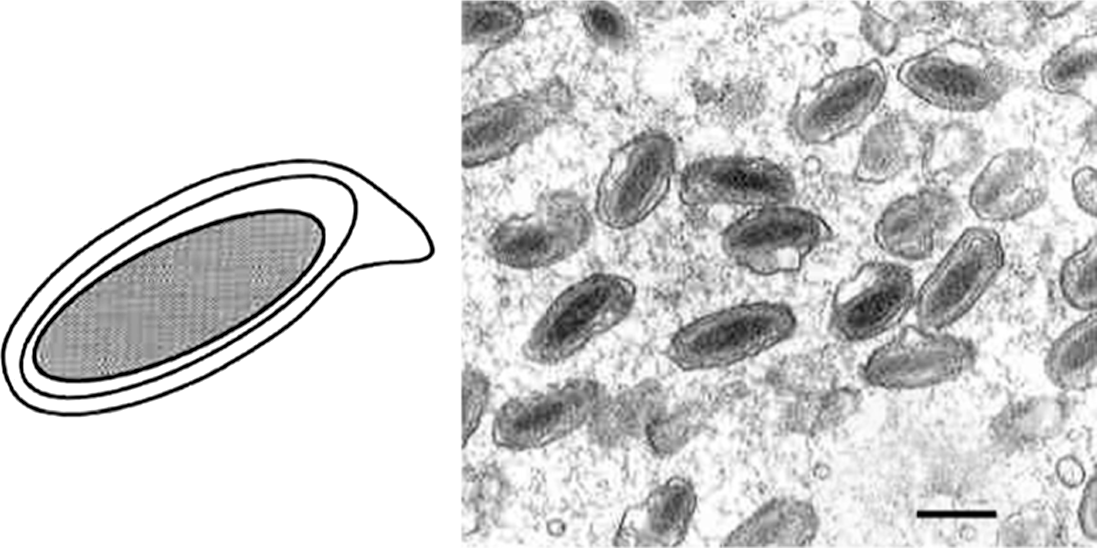
Figure 7 Undigested Campoletis sonorensis ichnovirus (CsIV) genome (left panel) with unique segment labels on right and nested segment labels on left. Colors indicate genes known to be associated with each segment. On left all segments encode cys-motif genes. Segments indicated in red are derived from larger segments (nested). Labels on right in this panel indicate unique segments that encode rep genes (black) and non-rep genes (e.g. P has similarity to NF- genes). Right panel illustrates unique (top) and nested segments (bottom) in their proviral (integrated-linear) and episomal forms (circular).
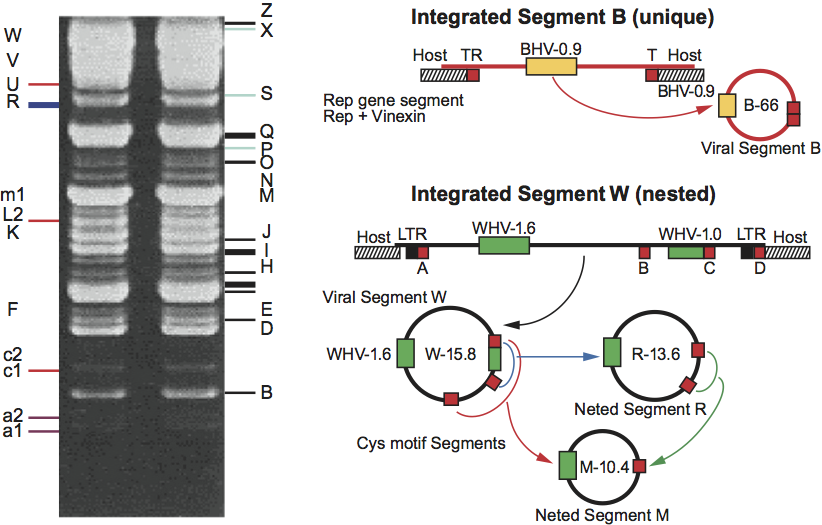
Figure 8 Bracovirus and ichnovirus phylogenetic tree illustrating that all members of the Polydnaviridae are associated with parasitoid wasps in the superfamily Ichneumonoidea. Subfamilies of the Braconidae that carry bracoviruses and subfamilies of the Ichneumonidae that carry ichnoviruses are shown. Parentheses indicate the approximate number of wasp species in each subfamily.