Family Podoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Distinguishing features
Virions have short, non-contractile tails about 20×8 nm. Heads are assembled first and tail parts are added to them sequentially. The classification of the family has recently been revised and is based on available proteomic data, linked to biological properties that include morphology, genome organization, mechanisms of DNA packaging and presence of DNA or RNA polymerase genes on the genome. Phages were clustered in groups using various clustering algorithms and careful review of available literature and experimental data. Analysis and biological interpretation of the molecular correlations among all tailed phages (Caudovirales) with known genome sequence, allowed the relationship between two phages to be summarized in a single correlation score (= the relative number of homologous proteins between two sequenced phages). Using a cut-off score of 40% homologous proteins between two phages, phages cluster correctly within existing genera. Higher level relationships (20% correlation) supported the introduction of subfamilies. Subfamilies emphasize commonalities between related genera and prevent excessive subdivision during classification.
Genus “BPP-1-like viruses”
Type species Bordetella phage BPP-1
Distinguishing features
The “BPP1-like viruses” include temperate viruses (infecting members of the Betaproteobacteria Bordetella [BPP-1, BMP-1 and BIP-1] and Burkholderia [BcepC6B]). They share the presence of genes specifying DNA polymerase I, a helicase and repressors.
BPP-1-like phages share some common gene products with the temperate VP2-related and “Epsilon15-like” phages. However, this homology is largely restricted to the proteins involved in morphogenesis and these phages have not therefore been grouped into a single subfamily.
Virion properties
Morphology
Phage BPP-1 particles have a 60 nm icosahedral capsid, a short tubular tail with a collar, and six tail fibers with bilobed globular ends (Figure 1).
Physicochemical and physical properties
No information available.
Nucleic acid
Linear doubled stranded DNA genomes of 42.4–42.7 kb with a G+C content of 65.2–65.4%.
Proteins
Genomes encode 46–49 proteins. The Bordetella phages code for a 38.0 kDa protein Brt (gp5) with a homology to RNA-directed DNA polymerases (reverse transcriptase) which plays a role in tropism.
Lipids
None known.
Carbohydrates
None known.
Genome organization and replication
The Bordetella phages integrate into host his-tRNA genes (attP TGGGGTGGCTGATG-GGACTCGAACCCA). The nature of the Burkholderia phage attP site is not known.
Biological properties
No information available.
Species demarcation criteria in the genus
The species have different hosts and only about 40% of the predicted gene products show proteomic correlation to other species. There is no genome-wide DNA homology between species.
List of species in the genus “BPP-1-like viruses”
| Bordetella phage BPP-1 |
|
|
| Bordetella phage BPP-1 | [AY029185=NC_005357] | (BPP-1) |
| Burhkolderia phage BcepC6B |
|
|
| Burhkolderia phage BcepC6B | [AY605181=NC_005887] | (BcepC6B) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed
List of other related viruses which may εbe members of the genus “BPP-1-like viruses” but have not been approved as species
None reported. BMP-1 was derived from BPP-1 through repeated passage on an alternative strain selecting for tropism switching. BPP-1 was isolated from a different lysogen of Bordetella bronchiseptica.
Genus “Epsilon15-like viruses”
Type species Salmonella phage epsilon15
Virion properties
Morphology
Heads are icosahedra with 60 hexamers and 11 pentamers, and measure about 70 nm in diameter in cryo-EM images. Tails are about 15 nm in length and there are six tailspikes (gp20) surrounding an external tail hub (17×14 nm; Figure 2).
Physicochemical and physical properties
No information available.
Nucleic acid
Genomes are about 39 kbp, linear and have a G+C content of 48–50%.
Proteins
Only the virions of ε15 have been studied in depth. They possess six structural proteins as shown by SDS-PAGE and mass spectrometry. These include 420 copies of the major capsid protein (gp7; 36.8 kDa, but maybe cleaved and cross-linked), 140 copies of the putative head stabilization protein gp10 (12.2 kDa), 12 copies of the portal protein (gp4; 61.7 kDa), putative hub-associated proteins gp15 (91 kda), gp16 (67.4 kDa) and gp17 (100 kDa); and, tailspike protein gp20 (115.7 kDa; possesses endorhamnosidase activity). Both ε15 and V10 carry O-antigen modification cassettes.
Lipids
None known.
Carbohydrates
None known.
Genome organization and replication
The genomes are linear, terminally redundant and circularly permuted with approximately 40 kbp (48–50% G+C). They encode 53 proteins, and no tRNAs. Simplistically speaking, the genes are arrayed in two transcriptional units, one on the negative strand involved in regulation and recombination, and the other on the positive strand, involved in packaging, morphogenesis, lysis and integration. The attachment sites of these phages are a subset of ATTGAGTGGGAATGATT which overlap the 3’ end of guaA. This site is located upstream of the integrase gene. Both phages carry lipopolysaccharide-modifying cassettes.
Biological properties
Phages are temperate and infect Salmonella and Escherichia coli strains. Distribution is unknown.
Species demarcation criteria in the genus
The species have different hosts and only about 40% of the predicted gene products show proteomic correlation to other species. There is no genome-wide DNA homology between species.
List of species in the genus “Epsilon15-like viruses”
| Salmonella phage epsilon15 |
|
|
| Salmonella phage ε15 | [AY150271=NC_004775] | (Eta15) |
| Escherichia phage PhiV10 |
|
|
| Escherichia phage V10 | [DQ126339=NC_007804] | (V10 or V10) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listedIt is quite likely that V10 is identical to phage V10 which is part of the O157 typing set. V10 and ε15 are related to a genomic island designated as PHIAP20 in E. coli APEC O1.
List of other related viruses which may be members of the genus “Epsilon15-like viruses” but have not been approved as species
None reported.
Genus “LUZ24-like viruses”
Type species Pseudomonas phage LUZ24
Virion properties
Morphology
Virions carry an icosahedral head with a 63 nm diameter and a short tail (12×8 nm). Decoration proteins protruding from the phage capsid can be distinguished (Figure 3).
Physicochemical and physical properties
No information available.
Nucleic acid
The “LUZ24-like viruses” contain linear dsDNA of around 45 kb. DNA of Pseudomonas phage LUZ24 was shown to have terminal direct repeats of 184 bp.
Proteins
Only a limited number of protein functions could be predicted by similarity searches. Using mass spectrometry, nine virion-associated proteins have been identified, delineating the structural region of LUZ24 from gene product 49 to gene product 65.
Lipids
None known.
Carbohydrates
None known.
Genome organization and replication
The genome of LUZ24 and PaP3 share an overall nucleotide sequence identity of 71%. Eight insertions/deletions are present and 88% of the encoded gene products are related. Type species LUZ24 encodes 68 proteins, 47 genes are arranged in rightward orientation while 21 are leftward. Two tRNA genes (tRNAAsn and tRNAPro) are present at the right end of the LUZ24 genome, compared to four in the genome of PaP3. LUZ24 shares its bidirectional genome organization with a vast number of virulent phages like cyanophages P60, Pf-WMP3 and Pf-WMP4, Roseophage SIO1 and vibriophage VpV262 in the subfamily Autographinae. These phage also share a strikingly conserved order of structural genes suggesting that these features are of very ancient origin.
Biological properties
Host range screenings on environmental and clinical P. aeruginosa strains revealed that LUZ24 lyses 36 out of 123 (29%) strains. Small and turbid plaques (1 mm) arise after infection of P. aeruginosa PAO1, although the phage extracted from such plaques appears not to differ from normal in latent period or burst size. Stable lysogenic P. aeruginosa clones could not be isolated. No phages could be induced, and no integrated LUZ24 DNA sequences could be demonstrated by PCR or restriction analysis. These data suggest that the phages are lytic.
Species demarcation criteria in the genus
The overall DNA sequence similarity between LUZ24 and PaP3 genomes is 71%; 88% of the encoded gene products are homologous.
List of species in the genus “LUZ24-like viruses”
| Pseudomonas phage LUZ24 |
|
|
| Pseudomonas phage LUZ24 | [AM910650=NC_010325] | (LUZ24) |
| Pseudomonas phage PaP3 |
|
|
| Pseudomonas phage PaP3 | [AY078382=NC_004466] | (PaP3) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “LUZ24-like viruses” but have not been approved as species
None reported.
Genus“N4-like viruses”
Type species Escherichia phage N4
Virion properties
Morphology
Virions have icosahedral heads about 70 nm in diameter and short tails 10 nm in length, with several short fibers originating from the junction between the head and tail (Figure 4).
Physicochemical and physical properties
Particle weight is about 84×106. Buoyant density in CsCl is 1.500 g cm-3. Lipids have not been detected in the established species.
Nucleic acid
Virions contain a single molecule of ds DNA of 70,153 bp of unique sequence flanked by 390–440 bp direct terminal repeats (variable lengths within the population). There are also short 3’ ssDNA extensions at both ends. G+C content is 41.3%.
Proteins
Virions contain at least 10 structural proteins. The major CP (ca. 500 molecules per phage) has a subunit mass of 44.0 kDa.
Lipids
Lipids (2.4%) have been reported in phage Sd, a putative member of this genus.
Carbohydrates
None reported.
Genome organization and replication
The physical map is linear, but a genetic linkage map has not been determined for technical reasons. There are 72 protein-coding genes identified which occupy 94% of the DNA sequence. Transcription of N4 DNA is carried out by sequential activity of three different RNA polymerases. Early transcription is performed by the viral RNA polymerase, a large (3500 aa) single subunit enzyme that is present in 1–2 copies in the virion and injected with the DNA during infection. Early proteins include a two-subunit RNA polymerase responsible for middle transcription. Middle proteins include a 98 kDa DNA polymerase and other DNA replication functions that replicate phage DNA in cooperation with some host functions. Late transcription is carried out by the host (E. coli) RNA polymerase. Early and middle genes occupy a contiguous block in the left half of the genome and are transcribed rightwards. Late genes occupy the right half of the genome and are transcribed leftwards. Other noteworthy genes include three tRNA genes (Asn, Thr, Pro), a gene encoding a dCTP deaminase, homologs of the rIIA and rIIB genes of phage T4, and a homolog of the gene 17 of temperate phage P22. Assembling particles form intracellular crystal-like arrays of phage heads.
Biological properties
Phages are virulent. Infection is lytic. Host range was originally thought to be restricted to enterobacteria (E. coli), but related Pseudomonas phages have been isolated recently.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus “N4-like viruses”
| Escherichia phage N4 |
|
|
| Escherichia coli bacteriophage N4 | [EF056009=NC_008720] | (N4) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “N4-like viruses” but have not been approved as species
| Silicibacter phage DSS3Phi2 | [FJ591093=NC_012697] | (DSS3Phi2) |
| Sulfitobacter phage EE36Phi1 | [FJ591094=NC_012696] | (EE36Phi1) |
| Pseudomonas phage LIT1 | [FN422399=NC_013692] | (LIT1) |
| Pseudomonas phage LUZ7 | [FN422398=NC_013691] | (LUZ7) |
| Enterobacteria phage PEV2 |
| (PEV2) |
| Enterobacteria phage Sd |
| (Sd) |
Genus “P22-like viruses”
Type species Enterobacteria phage P22
Distinguishing features
Virions have short tails which have six prominent tail spikes. DNA is circularly permuted, terminally redundant, and packaged from a pac site by a headful mechanism. Phages can carry out generalized transduction. The genetic map is circular.
Virion properties
Morphology
P22 virions have isometric icosahedral heads with 72 faintly visible capsomers (60 hexamers, 12 pentamers; T=7) and 59 to 63 nm (three-fold and five-fold axes, respectively) in diameter (Figure 5). Tails are 18 nm long and have six tail spikes.
Physicochemical and physical properties
Virion buoyant density in CsC1 is about 1.50 g cm-3 and S20,w is 500S. Infectivity is ether- and chloroform-resistant.
Nucleic acid
The P22 genome is linear double stranded DNA of 41,754 bp. The chromosome in virions ranges from 42.7 to 44.1 kbp, corresponds to about 55% of particle weight, and has a G+C content of 47%. Other members of the genus have genome sequences of 38–42 kbp with similar organization and similar transcriptional programs.
Proteins
P22 virions contain nine structural proteins: 415 copies of the major capsid protein (gp5; 46.7 kDa), 12 copies of the portal protein gp1 (82.7 kDa), two protein, gp4 (18.0 kDa) and gp10 (52.5 kDa), which are part of the hub, gp7 (23.4 kDa), gp16 (64.4 kDa) and gp20 (50.1 kDa) which are pilot/injection proteins, the tail needle protein gp26 (three copies, 24.7 kDa) and 18 copies of the tailspike/endorhamnosidase protein gp9 (71.9 kDa). Other members of the genus are similar.
Lipids
None known.
Carbohydrates
None known.
Genome organization and replication
The P22 genetic map is circularly permuted, has terminal repeats of about 1600 bp (3.8% of the genome), and comprises at least 65 genes. The genome sequence is partially (13.5%) similar to that of coliphage λDNA and common sequences are scattered across the right half of the genome. Other genus members have similar but different relationships with other “lambda-like viruses”. The integration system is dispensable for lytic growth. Transcription starts with regulatory genes and proceeds in three partly overlapping waves that are very similar to those of the lambda-like viruses. Replication starts at a single site and involves replication by a θ (theta) structure mechanism that switches at late times to a rolling-circle mechanism (σ, sigma replication). Progeny DNA is cut from concatemers at a pac site and packaged by a headful mechanism.
Antigenic properties
No group antigens are reported.
Biological properties
Phages are temperate and can carry out generalized transduction with lysogenic conversion ability. Members of the genus infect Enterobacteria (Escherichia, Salmonella and other Gammaproteobacteria) and have, under suitable conditions, very high (up to 500) burst sizes. Phage genomes integrate at specific sites in the bacterial genome and are UV-inducible.
Species demarcation criteria in the genus
Lack of DNA homology between species.
List of species in the genus “P22-like viruses”
| Enterobacteria phage P22 |
|
|
| Enterobacteria phage P22 | [BK000583=NC_002371] | (P22) |
| Salmonella phage ε34 | [EU570103=NC_011976] | (ε34) |
| Enterobacteria phage ST104 | [AB102868=NC_005841] | (ST104) |
| Salmonella phage HK620 |
|
|
| Salmonella phage HK620 | [AF335538=NC_002730] | (HK620) |
| Salmonella phage ST64T |
|
|
| Salmonella phage ST64T | [AY052766=NC_004348] | (ST64T) |
| Shigella phage Sf6 |
|
|
| Shigella phage Sf6 | [AF547987=NC_005344] | (Sf6) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “P22-like viruses” but have not been approved as species
| Aeromonas phage Aa-1 |
| (Aa-1) |
| Azotobacter phage A12 |
| (A12) |
| Enterobacteria phage L |
| (L) |
| Enterobacteria phage LP7 |
| (LP7) |
| Enterobacteria phage MG40 |
| (MG40) |
| Enterobacteria phage PSA78 |
| (PSA78) |
| Hyphomicrobium phage HyPhi30 |
| (HyPhi30) |
| Pseudomonas phage 525 |
| (525) |
| Vibrio phage O6N-72P |
| (06N-72P) |
Other highly likely members of this genus include phages: CUS-3 (CP000711), c341 (FJ000341=NC_013059), SE1 (DQ003260=NC_011802), SETP1, SETP8, SETP10, SETP14, SETP15, SETP16 and MG178.
Genus “Phieco32-like viruses”
Type species Enterobacteria phage Phieco32
Distinguishing features
A comparatively rare morphotype among the Podoviridae, PhiEco32 possesses an elongated head and a unique mechanism of transcription.
Virion properties
Morphology
This virus displays C3 type morphology with an elongated head (ca. 145×44 nm) and a short tail (ca. 13×8 nm) which may carry short kinked tail fibers (Figure 6).
Physicochemical and physical properties
Not known.
Nucleic acid
The linear double stranded DNA genome has 77,554 bp with 193 bp terminal direct repeats (42.3% G+C). It comprises 128 ORFs and a gene for arginyl-tRNA (codon: AGA).
Proteins
Ten virion proteins were identified by SDS-PAGE and mass spectrometry: gp8 (portal), gp11 (major capsid), gp14 and gp15 (tail fibres) and gp13, gp18-19, gp25-26 and gp58. The capsid protein may also exist as a larger frameshift derivative.
Lipids
None known.
Carbohydrates
None known.
Genome organization and replication
The phiEco32 genome encodes enzymes for nucleotide metabolism (gp34, deoxynucleoside monophosphate kinase; gp37, high-affinity ADP-ribose binding protein (Macro_Poa1p_like); gp64, thymidylate synthase; gp65 a thioredoxin-like protein, and gp72 a dCTP deaminase) and DNA replication (gp62, a DNA-binding protein; gp33, 5‘-3’ exonuclease; gp74, 3‘-5’ exonuclease; gp75, primase/helicase; gp62, DNA ligase and a Family A (pfam00476) DNA polymerase (gp53).
This phage genome possesses several sequences bearing strong sequence similarity to the consensus sigma-70 RNA polymerase promoters of the host bacterium (TTGACA-N15-17-TATAAT). It encodes an ECF-like sigma factor (gp36, 25.8 kDa) which is found associated with RNA polymerase (RNP) isolated from phage-infected cells, along with phage protein gp79 (9.5 kDa). The promoters recognized by this modified RNP are not known.
Biological properties
None reported.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus “Phieco32-like viruses”
| Enterobacteria phage Phieco32 |
|
|
| Enterobacteria phage PhiEco32 | [EU330206=NC_010324] | (phiEco32) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “Phieco32-like viruses” but have not been approved as species
Recently Salmonella phage 7-11 was sequenced and suggested as a member of this genus.
Subfamily Autographivirinae
Distinguishing features
The T7, SP6 and PhiKMV-like viruses have long been recognized as being related and are often termed members of a “supergroup”. Based on comparative proteomics data, this grouping was established as a subfamily. The origin of the name for this subfamily refers to the “auto-graphein” or “self-transcribing” phages which encode their own (single subunit) RNA polymerase, a common characteristic among the phages of this subfamily. More importantly, these phages share a common general genome organization, with genes encoded solely on the Watson strand.
Within this subfamily, distinctive features warrant a separation into different genera. Phages SP6 and K1-5 have been considered an “estranged subgroup of the T7-like viruses”. Also, the “phiKMV-like viruses” carry their single-subunit RNA polymerase gene adjacent to the structural protein region of the genome instead of in the early (host conversion) region, and they lack readily identifiable phage promoters. This suggests a different transcription scheme for “phiKMV-like viruses” compared to the “T7-like viruses”.
It is possible that the cyanophages P60, Syn5 and P-SSP7 represent one or more additional genera in this subfamily; however, a reevaluation of the P60 genome is necessary to perform an accurate analysis. Synechococcus phage syn5 and Prochlorococcus phage P-SSP7, share a correlation to P60 (36% and 35% respectively). These phage are therefore treated as unassigned species within the subfamily.
Bacteriophage SIO1 is related to this subfamily from an evolutionary perspective, but falls outside it because of the absence of a phage-encoded RNA polymerase and greater differences at the genome organization level. The same argument can be given for marine bacteriophage VpV262 infecting Vibrio parahaemolyticus. Genomic data indicates that an ancestral component of a T7 viral supergroup is conserved in the marine environment, but overall genomic differences and the lack of a phage-encoded RNA polymerase places this virus outside the subfamily.
Phage KSY1 does contain a phage-encoded RNA polymerase and a T7-like transcription scheme, but its overall morphology, genome size and proteome make-up does not warrant a classification within this subfamily.
Genus “PhiKMV-like viruses”
Type species Pseudomonas phage phiKMV
Distinguishing features
The members of this genus share no DNA sequence homology whatsoever to other members of the Autographivirinae. While the general genome organization is conserved, the CoreGenes algorithm only detects nine clear homologous gene products between φKMV and T7 (http://binf.gmu.edu:8080/CoreGenes2.0/).
Virion properties
Morphology
Phage morphology is conserved compared to “T7-like viruses” and consists of a short tail and an icosahedral capsid of about 60 nm in diameter. Preliminary cryo EM reconstructions of φKMV does suggest the presence of head-spike proteins associated with the main capsid proteins.
Physicochemical and physical properties
KMV virion Mr is about 48×106, buoyant density in CsCl is 1.50 g cm−3, and S20,w is about 510S. Infectivity is ether and chloroform resistant.
Nucleic acid
The double-stranded DNA genome varies around 42 kb in size, with direct terminal repeats (between 200 and 500 bp). The genome has few type II restriction endonuclease recognition sites but does contain site-specific nicks, also observed in Escherichia coli phage T5.
Proteins
The structural proteome of the type species KMV has been determined experimentally by mass spectrometry, as have several of its enzymes (DNA ligase, lysins). The structural (virion-associated) peptidoglycan hydrolase domain, located at the C-terminus of the predicted internal protein gp36, has a high refolding capacity and is thermoresistant. This property may be of relevance upon DNA ejection into the host cell, as proposed for phage T7 (folding carpenter’s rule model).
Lipids
None known.
Carbohydrates
None known.
Genome organization and replication
While the “T7-like viruses” and the “phiKMV-like viruses” share a common, general genome organization, typical of the Autographivirinae, individual protein homology is limited to a core set of proteins. In addition, KMV and LKA1 carry a single-subunit RNA polymerase gene adjacent to the structural genome region (instead of in the early region in “T7-like viruses”), and they lack conserved phage promoters. These features suggest major differences in the transcription scheme for “phiKMV-like viruses”.
Biological properties
Phages are highly virulent with a short infection cycle, and infection typically leads to the formation of large (ca. 5 mm), clear plaques. Similar to many Pseudomonas phages, infection by KMV, LKD16 is pili-dependent.
Species demarcation criteria in the genus
Phages KMV and LKA1 have similar genome organization and are similar in 45% of their gene products. Based on DNA sequence analysis, phages KMV, LKD16, kF77 and LUZ19 are clearly related. Although LKD16 is closely related to KMV and shows strong similarity in 90% of its predicted ORFs, differences in host range and infectivity parameters have been shown, albeit insufficient to warrant the establishment as different species. Partial/complete genome sequencing on bacteriophages PT2, PT5 and PS028, MMA1, MMA2, FMV, PBK, LUZ24 show these six phages are also close relatives of KMV.
List of species in the genus “PhiKMV-like viruses”
| Pseudomonas phage phiKMV |
|
|
| Pseudomonas phage KMV | [AJ505558=NC_005045] | (PhiKMV) |
| Pseudomonas phage LKA1 |
|
|
| Pseudomonas phage LKA1 | [AM265639=NC_009936] | (LKA1) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “PhiKMV-like viruses” but have not been approved as species
| Pseudomonas phage LKD16 | [AM265638=NC_009935] | (LKD16) |
| Pseudomonas phage phikF77 | [FN263372=NC_012418] | (PhikF77) |
| Pseudomonas phage phi-2 | [FN594518=NC_013638] | (Phi-2) |
| Pseudomonas phage PT2 | [EU236438=NC_011107] | (PT2) |
| Pseudomonas phage PT5 | [EU056923=NC_011105] | (PT5) |
| Pseudomonas phage LUZ19 | [AM910651=NC_010326] | (LUZ19) |
| Caulobacter phage Cd1 | [GU393987] | (Cd1) |
| Ralstonia phage RSB1 | [AB451219=NC_011201] | (RSB1) |
| Klebsiella phage KP34 | [GQ413938=NC_013649] | (KP34) |
| Vibrio phage VP93 | [FJ896200=NC_012662] | (VP93) |
Genus “SP6-like viruses”
Type species Enterobacteria phage SP6
Distinguishing features
This group of viruses is distinguished from other members of the Caudovirales by tailspike-like proteins possessing hydrolytic activities which are encoded at the extreme right end of the genome. Although there are substantial similarities to the T7-like viruses, there are key proteomic differences shown by CoreGenes analysis.
Virion properties
Morphology
Bacteriophages SP6 and K1-5 possess icosahedral heads (65–68 nm in diameter). The tail is short and stubby (like T7) and has been described as possessing an “irregular bushy structure”. Additional appendages are apparent, perhaps reflecting the cell envelope hydrolytic activities carried by both SP6 and K1-5 virions.
Physicochemical and physical properties
Infectivity is ether- and chloroform-resistant.
Nucleic acid
Genomes are, on average, 44.7 kbp (43769 bp for SP6), have a G+C content of approximately 47%, and are non-permuted and terminally redundant (the redundancies range from 174–288 bp).
Proteins
Structural proteins have been predicted based on the relationship to other phage in the subfamily Autographivirinae.
Lipids
None known.
Carbohydrates
None known.
Genome organization and replication
SP6-like viruses are not fundamentally different from the T7-like viruses in their gene order: RNA polymerase, helicase/primase, DNA polymerase, exonuclease, portal, scaffold, major capsid, tail, internal proteins, terminase subunits. The SP6-like viruses are recognized as a separate genus on the basis of CoreGenes analysis, showing several distinct differences. They lack T7gp2.5 (single-stranded DNA-binding protein) homologs; the ligase homolog (SP6 gp24) is located immediately upstream of the morphogenesis genes (in T7-like viruses gp1.3 is an early gene); and, the packaging proteins (SP6 gp39 and gp40) are contiguous on the genomes while in the T7-like viruses the holin-lysin complex is found between them (T7 gp18 & gp19). The DNA polymerase of SP6 (gp13) is significantly larger (849 amino acids) than that of T7 (gp5; 704 amino acids).
Biological properties
Infect Salmonella, Escherichia coli and Erwinia strains.
Species demarcation criteria in the genus
Species are separated by host range and supported by proteomic analyses.
List of species in the genus “SP6-like viruses”
| Enterobacteria phage SP6 |
|
|
| Enterobacteria phage SP6 | [AY288927=NC_004831] | (SP6) |
| Enterobacteria phage K1-5 |
|
|
| Enterobacteria phage K1-5 | [AY370674=NC_008152] | (K1-5) |
| Enterobacteria phage K1E |
|
|
| Enterobacteria phage K1E | [AM084415=NC_007637] | (KE1) |
| Enterobacteria phage K5 |
|
|
| Enterobacteria phage K5 |
| (K5) |
| Erwinia amylovora phage Era103 |
|
|
| Erwinia phage Era103 | [EF160123=NC_009014] | (Era103) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “SP6-like viruses” but have not been approved as species
None reported.
Genus“T7-like viruses”
Type species Enterobacteria phage T7
Distinguishing features
Medium-sized lytic phages, with nonpermuted terminally redundant DNA that codes for both DNA and RNA polymerases. Heads contain a unique eight-fold symmetric core structure. DNA is injected stepwise rather than all at once.
Virion properties
Morphology
T7 phage heads are icosahedra, measure about 60 nm in diameter, and consist of 72 capsomers (60 hexamers and 12 pentamers; T=7). Tails measure 17×8 nm and have six short fibers (Figure 7).
Physicochemical and physical properties
T7 virion Mr is about 48 ×106, buoyant density in CsCl is 1.50 g cm–3, and S20,w is about 510S. Infectivity is ether and chloroform resistant.
Nucleic acid
Genomes are about 40 kbp (39,936 bp for T7), corresponding to 50% of virion particle weight, have a G+C content of 50%, and are non-permuted and terminally redundant.
Proteins
Particles have at least nine structural proteins (13–150 kDa), including about 420 copies of the major capsid protein (in T7, 38 kDa).
Lipids
None known.
Carbohydrates
None known.
Genome organization and replication
The T7 genetic map is linear, non-permuted and terminally redundant, and comprises about 55 genes, several of which overlap. Related functions cluster together (Figure 8). The genome encodes a type B (E. coli Pol II) DNA polymerase and RNA polymerase. Infection results in shut-off of host syntheses and a breakdown of the host genome. The start of replication requires phage-encoded DNA and RNA polymerase and has multiple origins. Transcription proceeds in three waves. Only one strand is transcribed. Replication is bidirectional and produces concatemers by end-to-end joining of intermediate forms. Irregular polyheads are frequently observed. Packaged DNA is cut at fixed sites.
Biological properties
Phages are virulent and are specific for enterics and related Gram-negative bacteria.
Species demarcation criteria in the genus
Species differ by host range and, insofar as known, DNA sequence similarity.
List of species in the genus “T7-like viruses”
| Enterobacteria phage T7 |
|
|
| Enterobacteria phage T7 | [V01146=NC_001604] | (T7) |
| Kluyvera phage Kvp1 |
|
|
| Kluyvera phage Kvp1 | [FJ194439=NC_011534] | (Kvp1) |
| Pseudomonad phage gh-1 |
|
|
| Pseudomonas phage gh-1 | [AF493143=NC_004665] | (gh-1) |
| Vibrio phage VP4 | [DQ029335=NC_007149] | (VP4) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “T7-like viruses” but have not been approved as species
| Enterobacteria phage 285P | [GQ468526] | (285P) |
| Enterobacteria phage 13a | [EU734174=NC_011045] | (13a) |
| Enterobacteria phage EcoDS1 | [EU734172=NC_011042] | (EcoDS1) |
| Enterobacteria phage BA14 | [EU734171=NC_011040] | (BA14) |
| Enterobacteria phage T3 | [AY318471=NC_003298] | (T3) |
| Klebsiella phage KP32 | [GQ413937=NC_013647] | (KP32) |
| Klebsiella phage K11 | [EU734173=NC_011043] | (K11) |
| Morganella phage MmP1 | [EU652770=NC_011085] | (MnP1) |
| Salmonella phage phiSG-JL2 | [EU547803=NC_010807] | (phiSG-JL2) |
| Vibrio phage N4 | [FJ409640=NC_013651] | (N4) |
| Yersinia phage Yepe2 | [EU734170=NC_011038] | (Yepe2) |
| Enterobacteria phage H |
| (H) |
| Enterobacteria phage W31 |
| (W31) |
| Enterobacteria phage WPK |
| (WPK) |
| Enterobacteria phage PhiI |
| (PhiI) |
| Enterobacteria phage PhiII |
| (PhiII) |
| Enterobacteria phage BA14 |
| (Ba14) |
| Enterobacteria phage Phi1.2 |
| (Phi1.2) |
| Enterobacteria phage IV |
| (IV) |
| Enterobacteria phage K11 |
| (K11) |
| Enterobacteria phage PTB |
| (PTB) |
| Enterobacteria phage R |
| (R) |
| Enterobacteria phage Y |
| (Y) |
| Enterobacterial phage ViIII |
| (ViIII) |
| Escherichia phage K1F | [DQ111067=NC_007456] | (K1F) |
| Pseudomonas phage PhiPLS27 |
| (PhiPLS27) |
| Pseudomonas phage PhiPLS743 |
| (PhiPLS743) |
| Pseudomonas phage Psy9220 |
| (Psy9220) |
| Rhizobium phage 2 |
| (2) |
| Rhizobium phage S |
| (III) |
| Vibrio phage III |
| (III) |
| Yersinia phage PhiA1122 | [AY247822=NC_004777] | (PhiA1122) |
| Yersinia Berlin | [AM183667=NC_008694] | (Berlin) |
| Yersinia phage PhiYeO3-12 | [AJ251805=NC_001271] | (PhiYeO3-12) |
List of unassigned species in the subfamily Autographivirinae
| Prochlorococcus phage P-SSP7 |
|
|
| Prochlorococcus phage P-SSP7 | [AY939843=NC_006882] | (P-SSP7) |
| Synechococcus phage P60 |
|
|
| Synechococcus phage P60 | [AF189021=NC_003390] | (P60) |
| Synechococcus phage syn5 |
|
|
| Synechococcus phage syn5 | [EF372997=NC_009531] | (syn5) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
Subfamily Picovirinae
Distinguishing features
All these phages share unique properties, which differentiate them from other Podoviridae: a similar, special tail structure, their relatively small size and genome (with DNA with inverted terminal repeats), a similar gene number (20–29), a protein-primed DNA polymerase which, among phages, is found elsewhere only in the family Tectiviridae. Several genomic relationships to 29 shown by the CoreExtractor/CoreGenes analysis have previously been observed for phages 44AJHD, P68 and C1. These relatives of the 29-like phages were previously listed in the VIIIth ICTV Report. Here, 29 and its relatives are upgraded from a genus to a subfamily with two genera: the “Phi29-like viruses” and the “AHJD-like viruses”. Picovirinae refers to the small (Pico-) virion and genome sizes of the viruses within this subfamily, which represent the smallest tailed phages known.
The evolutionary link to the “Phi29-like viruses” is clearly present throughout the subfamily, both morphologically and molecularly, since all these phages also contain a type B polymerase, apart from other similar gene products and overall genome size. From this perspective, phages Actinomyces phage Av-1, Streptococcus phage Cp-1 are included within this subfamily but not assigned to a particular genus.
Mycoplasma phage P1 occupies a distinct and unclear position. Its genome has 11 structural genes, the same type of DNA polymerase as the other “phi29-like viruses”, and a genome size of only 11,660 bp. This needs confirmation since we may be observing a case of genome size reduction (as shown by Mycoplasma hosts themselves).
Genus “AHJD-like viruses”
Type species Staphylococcus phage 44AHJD
Distinguishing features
The “AHJD-like viruses” include Staphylococcus phages 44AHJD, P68, SAP-2 and 66, as well as Streptococcus phage C1. Evidence that distinguishes them from “phi29-like viruses” comes from their genome analysis which reveals that the 44AHJD and P68 lysis genes (amidases) are located within the morphogenesis genes rather than downstream, as is observed with 29. Furthermore, these phages lack a classical holin-lysin cassette. Lastly, the gene for the major capsid protein (gp8) of 29 and its close relatives is located in the left third of the genome, while the analogous genes in 44AHJD and P68 (gp20) are near the right end of the genome.
Virion properties
Morphology
Isometric phages, with short, non-contractile tails and a pre-neck appendage.
Physicochemical and physical properties
No information available.
Nucleic acid
These dsDNA viruses have genomes of between 16 and 19 kbp. The terminal DNA fragments of P68 DNA and P68 DNA protein complex suggest the presence of a terminal protein at either DNA end.
Proteins
These phages encode between 20 (phages C1 and SAP-2) and 27 (phage 66) proteins.
Lipids
None known.
Carbohydrates
None known.
Genome organization and replication
Phage 44AHJD and phage P68 genomes are predicted to contain 21 and 22 ORFs, respectively. Metabolism-related genes are encoded on the Watson strand of the genome, whereas genes encoding structural proteins are located on the Crick strand, separated by a bidirectional terminator. Evidence for the presence of a terminal protein in P68 has been published.
Biological properties
Phages 44AHJD, P68, phage 66 and SAP-2 are lytic phages infecting Staphylococcus aureus. Phage C1 infects Streptococcus.
Species demarcation criteria in the genus
The presence of unique genes.
List of species in the genus “AHJD-like viruses”
| Staphylococcus phage 44AHJD |
|
|
| Staphylococcus phage 44AHJD | [AF513032=NC_004678] | (44AHJD) |
| Staphylococcus phage phiP68 | [AF513033=NC_004679] | (phiP68) |
| Staphylococcus phage 66 | [AY954949=NC_007046] | (66) |
| Staphylococcus phage SAP-2 | [X99260=NC_004165] | (SAP2) |
| Streptococcus phage C1 |
|
|
| Streptococcus phage C1 | [AY212251=NC_004814] | (C1) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “AHJD-like viruses” but have not been approved as species
None reported.
Genus “Phi29-like viruses”
Type species Bacillus phage phi29
Virion properties
Morphology
Heads are prolate icosahedra (T=3 with 30 hexamers and 11 pentamers) and measure about 54×42 nm. Some members, including Bacillus phage 29, have about 55 fibers on the head (Figure 9). Tails measure 46×8 nm; have a distal thickening, and a collar with 12 appendages.
Physicochemical and physical properties
29 virion Mr is 29×107, buoyant density in CsC1 is 1.46 g cm–3, and S20,w is 254S. Infectivity is chloroform-resistant.
Nucleic acid
Genomes are 16–20 kbp, correspond to about 50% of particle weight, and have a G+C content of 35–38%.
Proteins
Virions have nine structural proteins (13-86 kDa), including 235 copies of the major capsid protein (49.6 kDa in 29 and 42 kDa in Cp-1). Type B (E. coli Pol II) DNA polymerase is coded for.
Lipids
None known.
Carbohydrates
None known.
Genome organization and replication
The genetic map is linear and includes 20–29 ORFs, one of which codes for a type B (E. coli Pol II) DNA polymerase. Genomes are non-permuted and have inverted terminal repeats from 6 to 8 bp (Phi29) to 230–240 bp (Cp-1). Both 5’ ends are covalently linked to a terminal protein. Infecting DNA does not circularize. Transcription proceeds in two waves. Early genes are transcribed from right to left on the standard map (except in Cp-1 where some are from left to right); late genes are transcribed from left to right. Replication is primed by the terminal protein and starts at both DNA ends, and proceeds by strand displacement. The terminal protein is essential in DNA packaging. The packaging substrate is non-concatemeric DNA. Packaging requires phage-encoded RNA.
Antigenic properties
No group antigens are reported.
Biological properties
Phages are virulent and infect Gram-positive bacteria with low G+C contents. Distribution is worldwide.
Species demarcation criteria in the genus
All species differ in host range. Phages 29 and GA-1 differ in serological properties and protein molecular weights. Phage 29, PZA, GA-1, B103, 44AHJD, P68, C1 and Cp-1 DNAs differ in DNA sequence and in the length of inverted terminal repeats (6–8 to 236–247 bp). In addition, phages 29 and Cp-1 have opposite directions for transcription of early genes at the left end of the chromosome.
List of species in the genus “Phi29-like viruses”
| Bacillus phage phi29 |
|
|
| Bacillus phage 29 | [EU771092=NC_011048] | (Phi29) |
| Bacillus phage GA-1 |
|
|
| Bacillus phage GA-1 | [X96987=NC_002649] | (GA-1) |
| Bacillus phage B103 |
|
|
| Bacillus phage B103 | [X99260=NC_004165] | (B103) |
| Kurthia phage 6 |
|
|
| Kurthia phage 6 |
| (K6) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “Phi29-like viruses” but have not been approved as species
| Bacillus phage Nf | [EU622808] | (Nf) |
| Bacillus phage BS32 | [B32RGHSEQ] | (BS32) |
| Bacillus phage Phi15 |
| (Phi15) |
| Bacillus phage AR13 |
| (AR13) |
| Bacillus phage MY2 |
| (MY2) |
| Bacillus phage M2 |
| (M2) |
| Bacillus phage N Phi |
| (N Phi) |
| Bacillus phage SF5 |
| (SF5) |
| Kurthia phage 7 |
| (K7) |
| Bacillus phage PZA | [9626384] | (PZA) |
About 45 additional, poorly characterized phages may belong to this genus.
List of unassigned species in the subfamily Picovirinae
| Actinomyces phage Av-1 |
|
|
| Actinomyces phage Av-1 | [DQ123818=NC_009643] | (Av-1) |
| Mycoplasma phage P1 |
|
|
| Mycoplasma phage P1 | [AF246223=NC_002515] | (P1) |
| Streptococcus phage Cp-1 |
|
|
| Streptococcus phage Cp-1 | [Z47794=NC_001825] | (Cp-1) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of unassigned species in the family Podoviridae
| Endosymbiont phage APSE-1 |
|
|
| Endosymbiont phage APSE-1 | [AF157835=NC_000935] | (APSE-1) |
| Lactococcus phage KSY1 |
|
|
| Lactococcus phage KSY1 | [DQ535032=NC_009817] | (KSY1) |
| Phormidium phage Pf-WMP3 |
|
|
| Phormidium phage Pf-WMP3 | [EF537008=NC_009551] | (Pf-WMP3) |
| Phormidium phage Pf-WMP4 |
|
|
| Phormidium phage Pf-WMP4 | [DQ875742=NC_008367] | (Pf-WMP4) |
| Pseudomonas phage 119X |
|
|
| Pseudomonas phage 119X | [DQ163914=NC_007807] | (119X) |
| Pseudomonas phage F116 |
|
|
| Pseudomonas phage F116 | [AY625898=NC_006552] | (F116) |
| Roseobacter phage SIO1 |
|
|
| Roseobacter phage SIO1 | [AF189021=NC_002519] | (SIO1) |
| Vibrio phage VpV262 |
|
|
| Vibrio phage VpV262 | [AY095314=NC_003907] | (VpV262) |
Species names are in italic script; strain names are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the family Podoviridae but have not been approved as species
| Acinetobacter phage A36 |
| (A36) |
| Bacillus phage PhiBa1 |
| (PhiBa1) |
| Brucella phage Tb |
| (Tb) |
| Clostridium phage HM2 |
| (HM2) |
| Coryneforms phage 7/26 |
| (7/26) |
| Coryneforms phage AN25S-1 |
| (AN25S-1) |
| Cyanobacteria phage A-4(L) |
| (A-4(L) |
| Cyanobacteria phage AC-1 |
| (AC-1) |
| Cyanobacteria phage LPP-1 |
| (LPP-1) |
| Cyanobacteria phage SM-1 |
| (SM-1) |
| Enterobacteria phage 7-11 | [HM997019] | (7-11) |
| Enterobacteria phage 7480b |
| (7480b) |
| Enterobacteria phage Esc-7-11 |
| (Esc-7-11) |
| Enterobacteria phage W8 |
| (W8) |
| Lactococcus phage PO34 |
| (PO34) |
| Mollicutes phage C3 |
| (C3) |
| Mollicutes phage L3 |
| (L3) |
| Mycobacterium phage Phi17 |
| (Phi17) |
| Pasteurella phage 22 |
| (22) |
| Rhizobium phage Phi2042 |
| (Phi2042) |
| Streptococcus phage 182 |
| (182) |
| Streptococcus phage 2BV |
| (2BV) |
| Streptococcus phage Cvir | [AF33467] | (Cvir) |
| Streptococcus phage H39 |
| (H39) |
| Vibrio phage 4996 |
| (4996) |
| Vibrio phage I |
| (I) |
| Vibrio phage OX N-100P |
| (OXN-100P) |
| Xanthomonas phage RR66 |
| (RR66) |
Phylogenetic relationships within the family
No information available.
Similarity with other taxa
“Phi29-like viruses“, adenoviruses and tectiviruses have proteins linked to DNA ends and protein-primed replication, and code for type B (E. coli Pol II) DNA polymerase. See “Similarity with other taxa” in the Caudovirales description for further details.
Further reading
Angly F, Youle M, Nosrat B, Srinagesh S, Rodriguez-Brito B, McNairnie P, Deyanat-Yazdi G, Breitbart M, Rohwer F. 2009. Genomic analysis of multiple Roseophage SIO1 strains. Environ Microbiol. 11:2863-2873.
Casjens, S., D. Winn-Stapley, E. Gilcrease, R. Moreno, C. Kühlewein, J. E. Chua, P. A. Manning, W. Inwood & A. J. Clark, (2004) The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. J.Mol. Biol. 339: 379-394.
Clark AJ, Inwood W, Cloutier T, Dhillon TS. (2001). Nucleotide sequence of coliphage HK620 and the evolution of lambdoid phages. J Mol Biol. 311:657-79.
Ceyssens PJ, Hertveldt K, Ackermann HW, Noben JP, Demeke M, Volckaert G, Lavigne R. (2004). The intron-containing genome of the lytic Pseudomonas phage LUZ24 resembles the temperate phage PaP3. Virology. 377:233-238.
Ceyssens PJ, Lavigne R, Mattheus W, Chibeu A, Hertveldt K, Mast J, Robben J, Volckaert G. 2006. Genomic analysis of Pseudomonas aeruginosa phages LKD16 and LKA1: establishment of the φKMV subgroup within the T7 supergroup. J Bacteriol. 188:6924-6931.
Ceyssens PJ, Brabban A, Rogge L, Lewis MS, Pickard D, Goulding D, Dougan G, Noben JP, Kropinski A, Kutter E, Lavigne R. (2010). Molecular and physiological analysis of three Pseudomonas aeruginosa phages belonging to the "N4-like viruses". Virology. 405:26-30.
Chen F, Lu J. 2002.Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl Environ Microbiol. 68:2589-9254.
Chopin A, Deveau H, Ehrlich SD, Moineau S, Chopin MC. (2007). KSY1, a lactococcal phage with a T7-like transcription. Virology. 365:1-9.
Dobbins AT, George M Jr, Basham DA, Ford ME, Houtz JM, Pedulla ML, Lawrence JG, Hatfull GF, Hendrix RW. 2004. Complete genomic sequence of the virulent Salmonella bacteriophage SP6. J Bacteriol. 186:1933-1944.
Dunn, J. and Studier, W. (1983). Complete nucleotide sequence of bacteriophage T7. J. Mol. Biol., 166, 477-535.
Hardies SC, Comeau AM, Serwer P, Suttle CA. 2003. The complete sequence of marine bacteriophage VpV262 infecting Vibrio parahaemolyticus indicates that an ancestral component of a T7 viral supergroup is widespread in the marine environment. Virology. 310:359-371.
Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. 2008. Backbone structure of the infectious ε15 virus capsid revealed by electron cryomicroscopy. Nature. 451:1130-1134.
Kiino, D.R. and Rothman-Denes, L.B. (1988). Bacteriophage N4. In: The Bacteriophages, vol. I, (R. Calendar, ed), pp 475-494. Plenum Press, New York.
King, M. R., R. P. Vimr, S. M. Steenbergen, L. Spanjaard, G. Plunkett, 3rd, F. R. Blattner & E. R. Vimr, (2007) Escherichia coli K1-specific bacteriophage CUS-3 distribution and function in phase-variable capsular polysialic acid O acetylation. J Bacteriol 189: 6447-6456
Kropinski AM, Sulakvelidze A, Konczy P, Poppe C. 2007. Salmonella phages and prophages-genomics and practical aspects. Methods Mol Biol. 394:133-75.
Kropinski AM, Kovalyova IV, Billington SJ, Patrick AN, Butts BD, Guichard JA, Pitcher TJ, Guthrie CC, Sydlaske AD, Barnhill LM, Havens KA, Day KR, Falk DR, McConnell MR. 2007. The genome of ε15, a serotype-converting, Group E1 Salmonella enterica-specific bacteriophage. Virology. 369:234-44.
Kropinski AM, Lingohr EJ, Ackermann HW. (2010). The genome sequence of enterobacterial phage 7-11, which possesses an unusually elongated head. Arch Virol. Nov 24
Lavigne R, Seto D, Mahadevan P, Ackermann HW, Kropinski AM. (2009). Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res Microbiol. 159:406-414.
Leiman PG, Battisti AJ, Bowman VD, Stummeyer K, Mühlenhoff M, Gerardy-Schahn R, Scholl D, Molineux IJ. 2007. The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. J Mol Biol. 371:836-849.
Liu, M., Gingery, M., Doulatov, S.R., Liu, Y., Hodes, A., Baker, S., Davis, P., Simmonds, M., Churcher, C., Mungall, K., Quail, M.A., Preston, A., Harvill, E.T., Maskell, D.J., Eiserling, F.A., Parkhill, J. and Miller, J.F. (2004). Genomic and genetic analysis of Bordetella bacteriophages encoding reverse transcriptase-mediated tropism-switching cassettes. J Bacteriol. 186:1503-1517.
Martín, A.C., López, R. and García, P. (1996). Analysis of the complete nucleotide sequence and functional organization of the genome of Streptococcus pneumoniae bacteriophage Cp-1. J. Virol., 70, 3678-3687.
Meijer, W.J., Horcajadas, J.A. and Salas, M. (2001). φ29 family of phages. Microbiol. Mol. Biol. Rev., 65, 261-287.
Nelson, D., Schuch, R., Zhu. S., Tscherne, D.M. and Fischetti, V.A. (2003). Genomic sequence of C1, the first streptococcal phage. J. Bacteriol., 185, 3325-32.
Pajunen, M.I., Kiljunen, S.J., Söderholm, M. and Skurnik, M. (2001). Complete sequence of the lytic bacteriophage φY3O3-12 of Yersinia enterocolitica serotype O:3. J. Bacteriol., 183, 1928-1937.
Pajunen, M.I., Elizondo, M.R., Skurnik, M., Kieleczawa, J. and Molineux, I.J. (2003). Complete nucleotide sequence and likely recombinatorial origin of bacteriophage T3. J. Mol. Biol., 319, 1115-1132.
Parent, K. N., R. Khayat, L. H. Tu, M. M. Suhanovsky, J. R. Cortines, C. M. Teschke, J. E. Johnson & T. S. Baker, (2010) P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks. Structure 18: 390-401.
Pedulla, M.L., Ford, M.E., Karthikeyan, T., Houtz, J.M., Hendrix, R.W., Hatfull, G.F., Poteete, A.R., Gilcrease, E.B., Winn-Stapley, D.A. and Casjens, S.R. (2003). Corrected sequence of the bacteriophage P22 genome. J. Bacteriol., 185, 1475-1477.
Pencenkova, T. and Paces, V. (1999). Molecular phylogeny of φ29-like phages and their evolutionary relatedness to other protein-primed replicating phages and other phages hosted by Gram-positive bacteria. J. Mol. Evol., 48, 197-208.
Pencenkova, T., Benes, V., Paces, J., Vlcek, C. and Paces, V. (1997). Bacteriophage B103: complete DNA sequence of its genome and relationship to other Bacillus phages. Gene, 199, 157-163.
Perry LL, SanMiguel P, Minocha U, Terekhov AI, Shroyer ML, Farris LA, Bright N, Reuhs BL, Applegate BM. 2009. Sequence analysis of Escherichia coli O157:H7 bacteriophage phiV10 and identification of a phage-encoded immunity protein that modifies the O157 antigen. FEMS Microbiol Lett. 292:182-186.
Rothman-Denes, L.B. (1995). DNA supercoiling, DNA hairpins and single-stranded DNA binding proteins in bacteriophage N4 transcription regulation. Sem. Virol., 6, 15-23.
Savalia D, Westblade LF, Goel M, Florens L, Kemp P, Akulenko N, Pavlova O, Padovan JC, Chait BT, Washburn MP, Ackermann HW, Mushegian A, Gabisonia T, Molineux I, Severinov K. (2008) Genomic and proteomic analysis of phiEco32, a novel Escherichia coli bacteriophage. J Mol Biol. 377:774-89.
Scholl D, Adhya S, Merril CR. 2002. Bacteriophage SP6 is closely related to phages K1-5, K5, and K1E but encodes a tail protein very similar to that of the distantly related P22. J Bacteriol. 184:2833-2836.
Scholl D, Kieleczawa J, Kemp P, Rush J, Richardson CC, Merril C, Adhya S, Molineux IJ. 2004. Genomic analysis of bacteriophages SP6 and K1-5, an estranged subgroup of the T7 supergroup. J Mol Biol. 335(5):1151-71. PubMed PMID: 14729334.
Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3:e144
Summer, E.J., Gonzalez, C.F., Bomer, M., Carlile, T., Morrison, W., Embry, A., Kucherka, A.M., Lee, J., Mebane, L., Morrison, W.C., Mark, L., King, M.D., LiPuma, M.J., Vidaver, A.K. and Young, R. (2006) Divergence and mosaicism among virulent soil phages of the Burkholderia cepacia complex. J. Bacteriology 188:255-268
Tanaka K, Nishimori K, Makino S, Nishimori T, Kanno T, Ishihara R, Sameshima T, Akiba M, Nakazawa M, Yokomizo Y, Uchida I. (2004). Molecular characterization of a prophage of Salmonella enterica serotype Typhimurium DT104. J Clin Microbiol. 42:1807-12.
Tu, AH, Voelker, LL, Shen, X and Dybvig, K. (2001) Complete nucleotide sequence of the mycoplasma virus P1 genome. Plasmid 45, 122-126.
Villafane R, Zayas M, Gilcrease EB, Kropinski AM, Casjens SR. (2008) Genomic analysis of bacteriophage ε34 of Salmonella enterica serovar Anatum (15+). BMC Microbiol. 8:227.
Vander Byl C, Kropinski AM. (2000) Sequence of the genome of Salmonella bacteriophage P22. J Bacteriol. 182:6472-81.
Vybiral, D., Takác, M., Loessner, M., Witte, A., von Ahsen, U. and Bläsi, U. (2003). Complete nucleotide sequence and molecuar characterization of two lytic Staphylococcus aureus phages: 44HJD and P68. FEMS Microbiol. Lett., 219, 275-283.
Zhang, Z., Greene, B., Thuman-Commike, P.A., Jakana, J. Prevelige, P.E., Jr., King, J. And Chiu, W. (2000). Visualization of the maturation transition in bacteriophage P22 by electron cryomicroscopy. J. Mol. Biol., 297, 615-626.
Contributed by
Lavigne, R. and Kropinski, A.M.
Figures
Figures
Figure 1 Phage BPP-1 negatively stained with 1% uranyl acetate, and an interpretive diagram.
(From Liu et al. (2004). J. Bacteriol., 186, 1503-1517; with permission.)
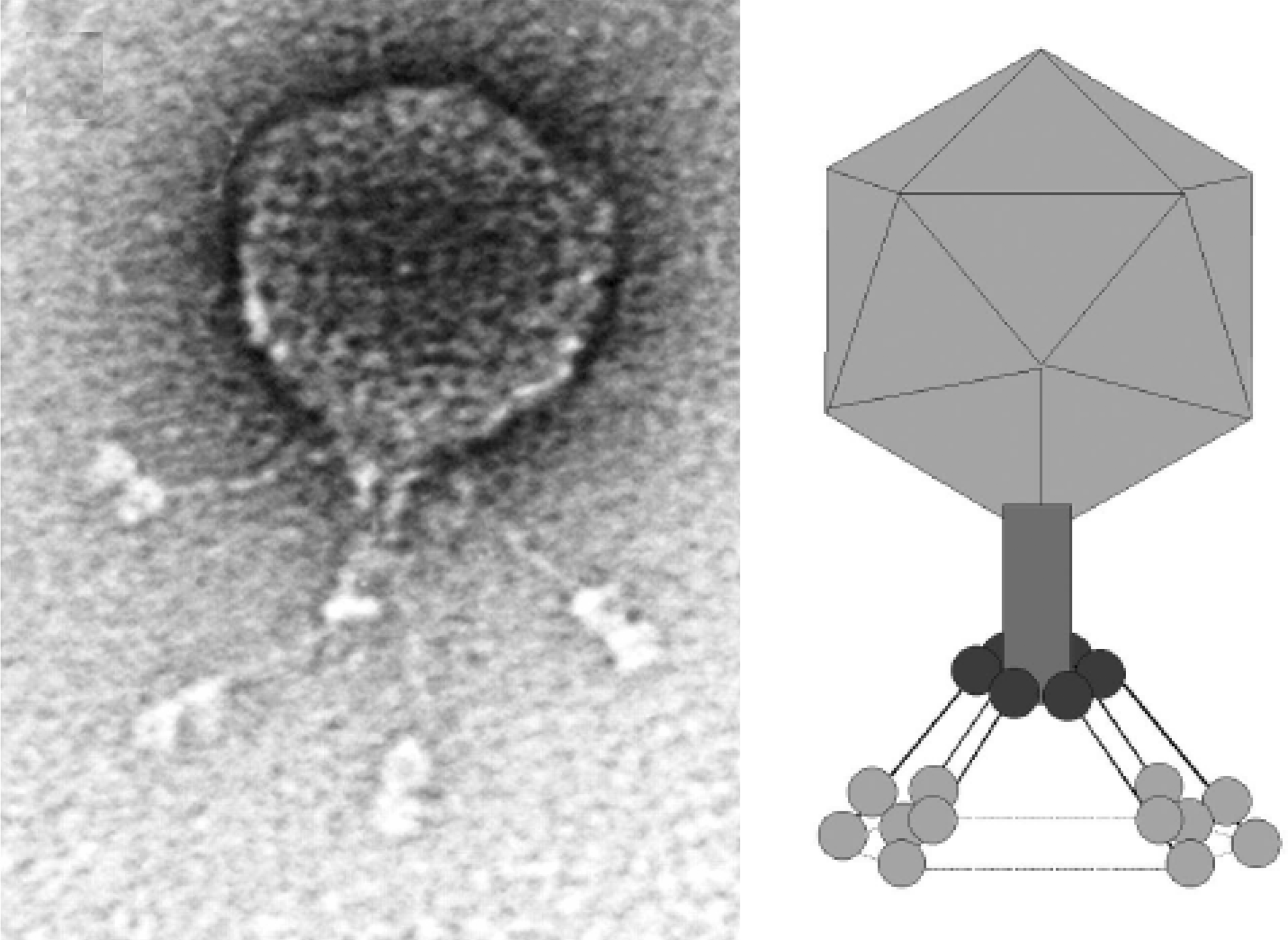
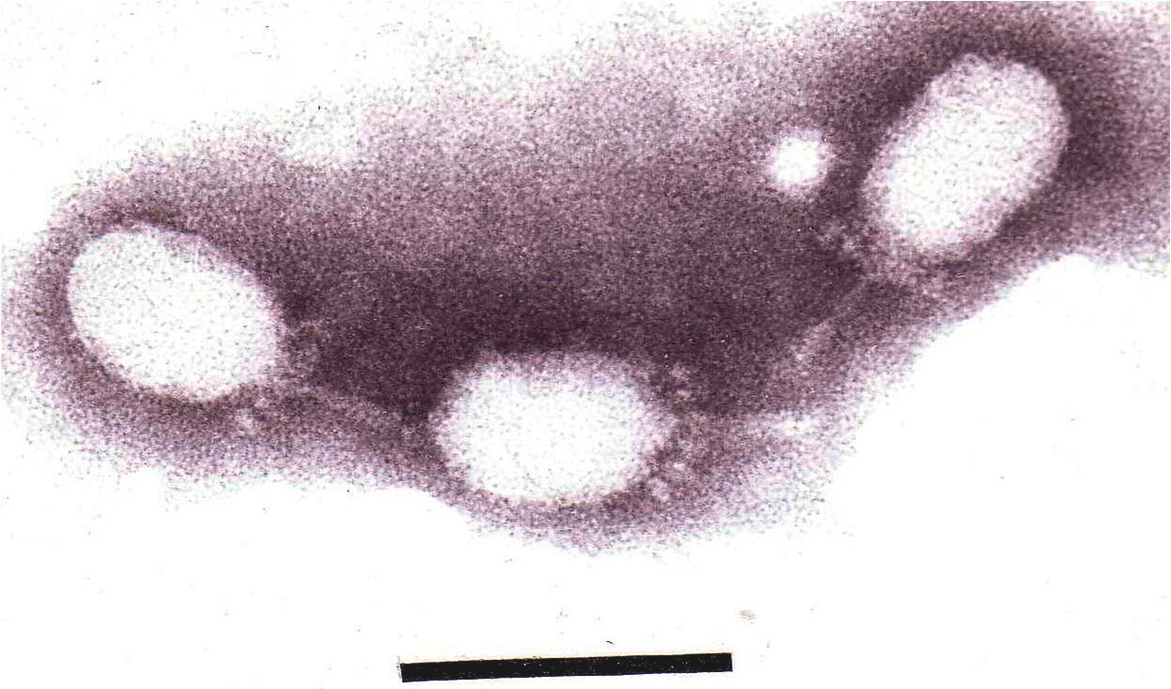
Figure 2 Electron micrograph of Salmonella phage 15 stained with phosphotungstic acid. The bar represents 100nm.
(Courtesy of Wen Jiang and Wah Chiu.)
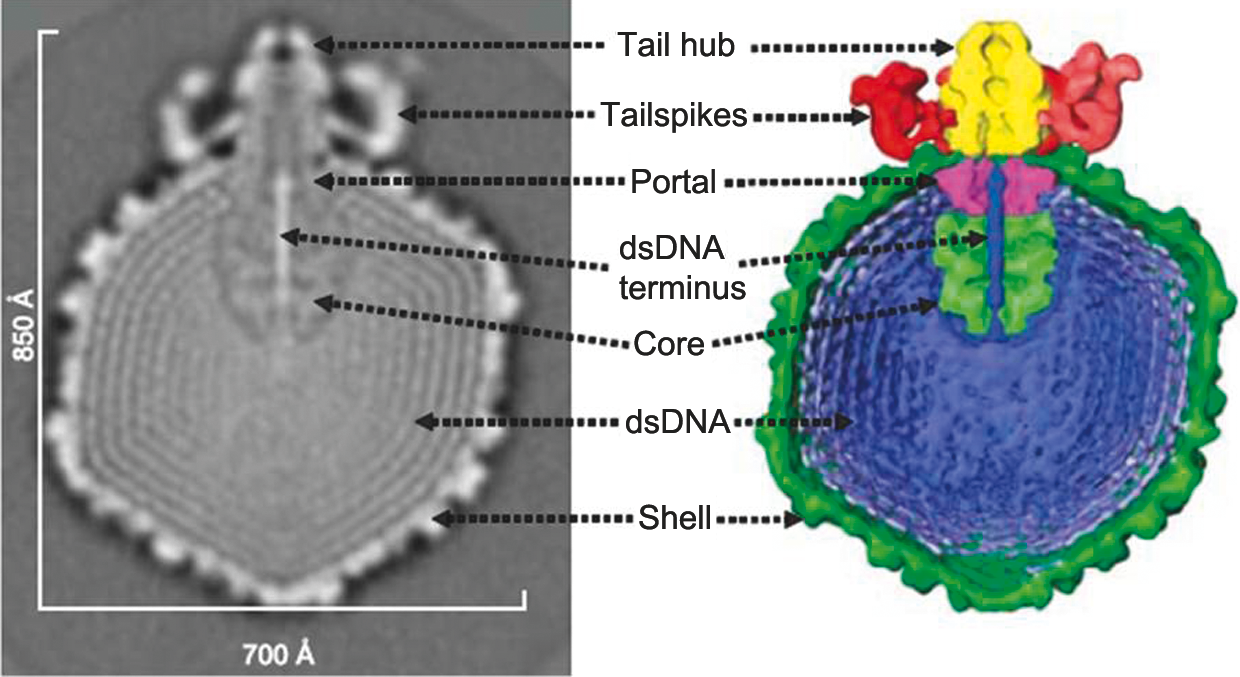
Figure 3 Electron micrograph of Pseudomonas phage LUZ24.
(Courtesy Hans-Wolfgang Ackermann.)
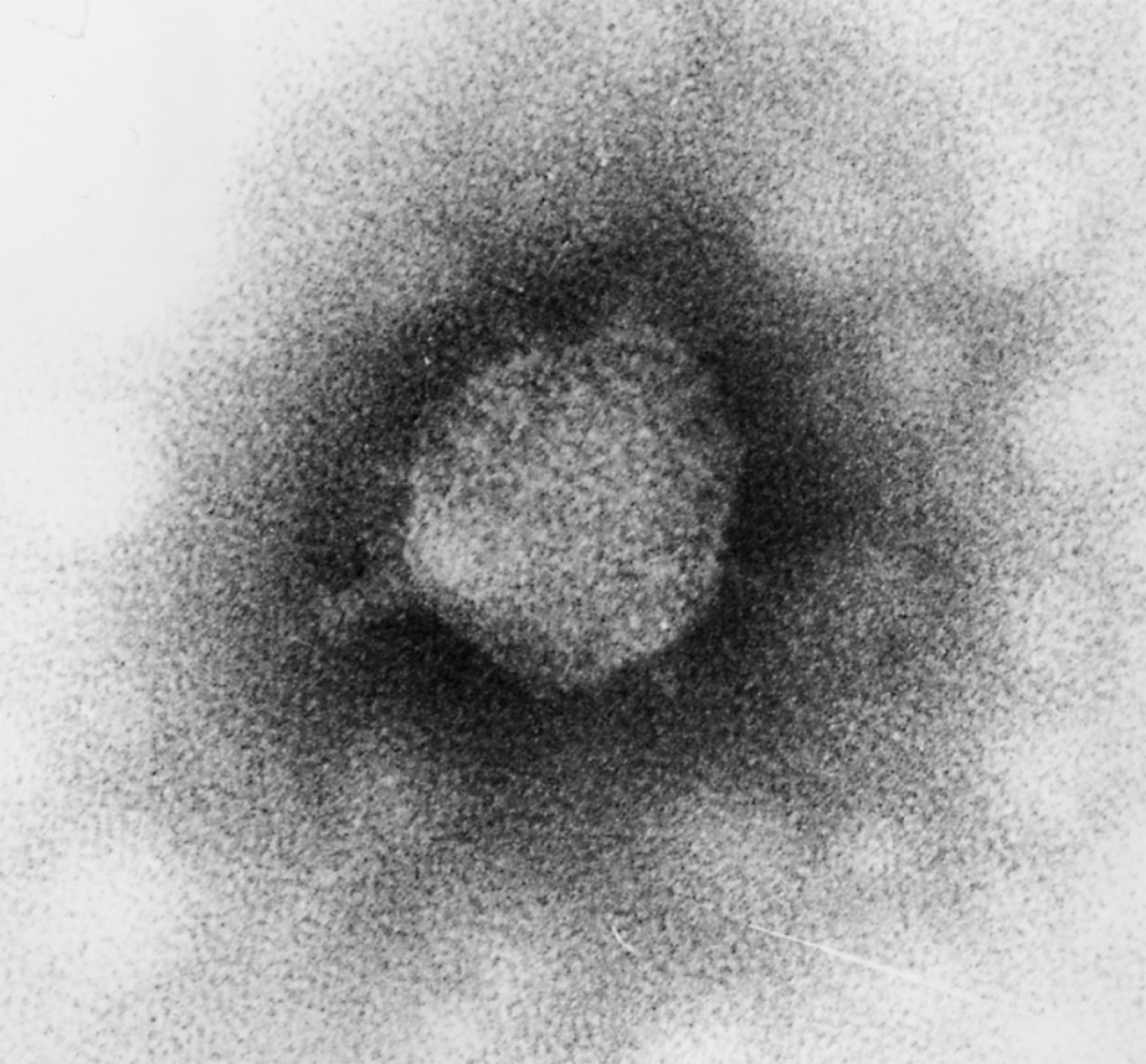
Figure 4 Electron micrograph of N4 stained with phosphotungstate. The bar represents 100 nm.
(Courtesy Hans-Wolfgang Ackermann.)
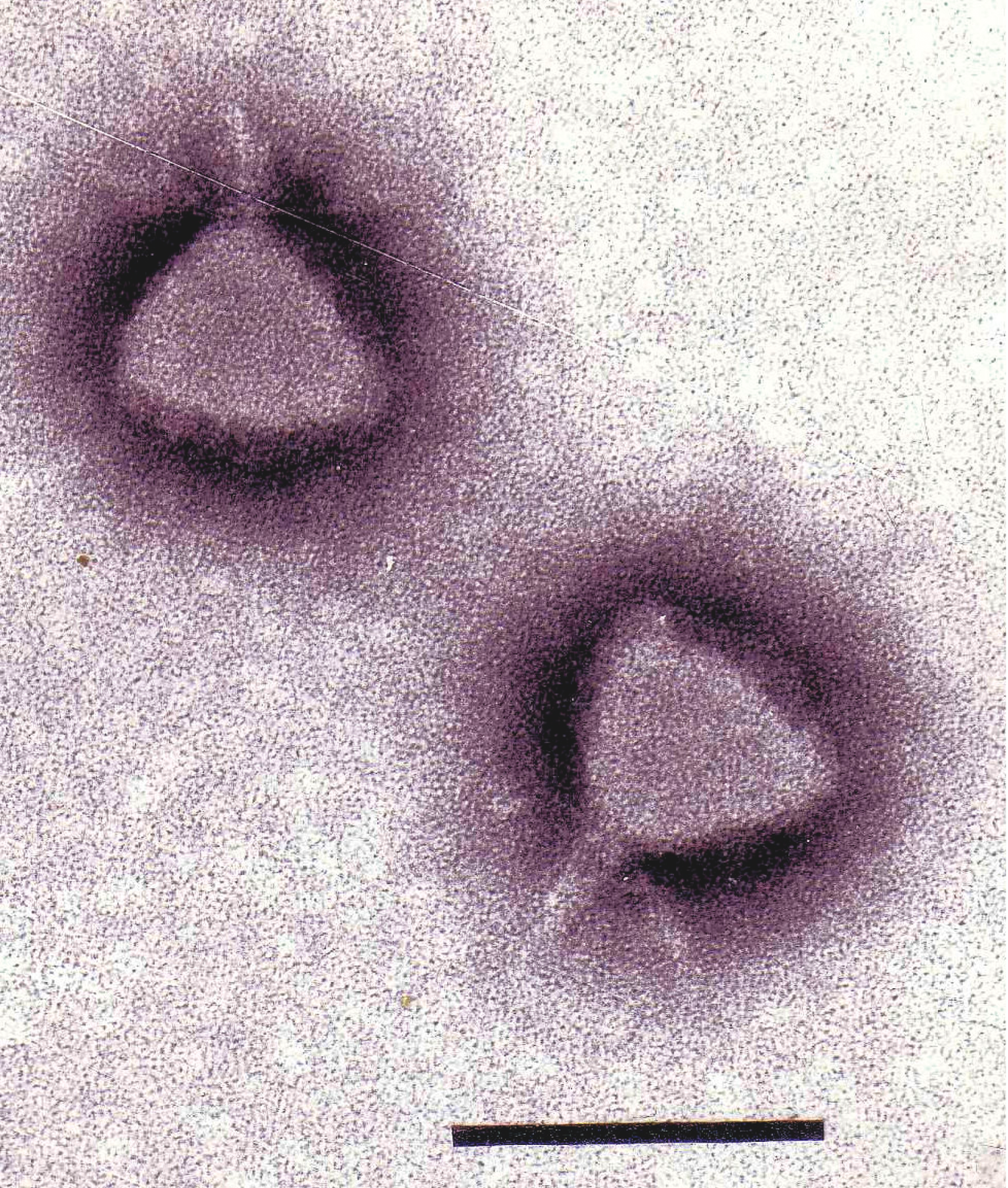
Figure 5 Phage P22 stained with uranyl acetate. The bar represents 100 nm bar.
(Courtesy Sherwood Casjens.)
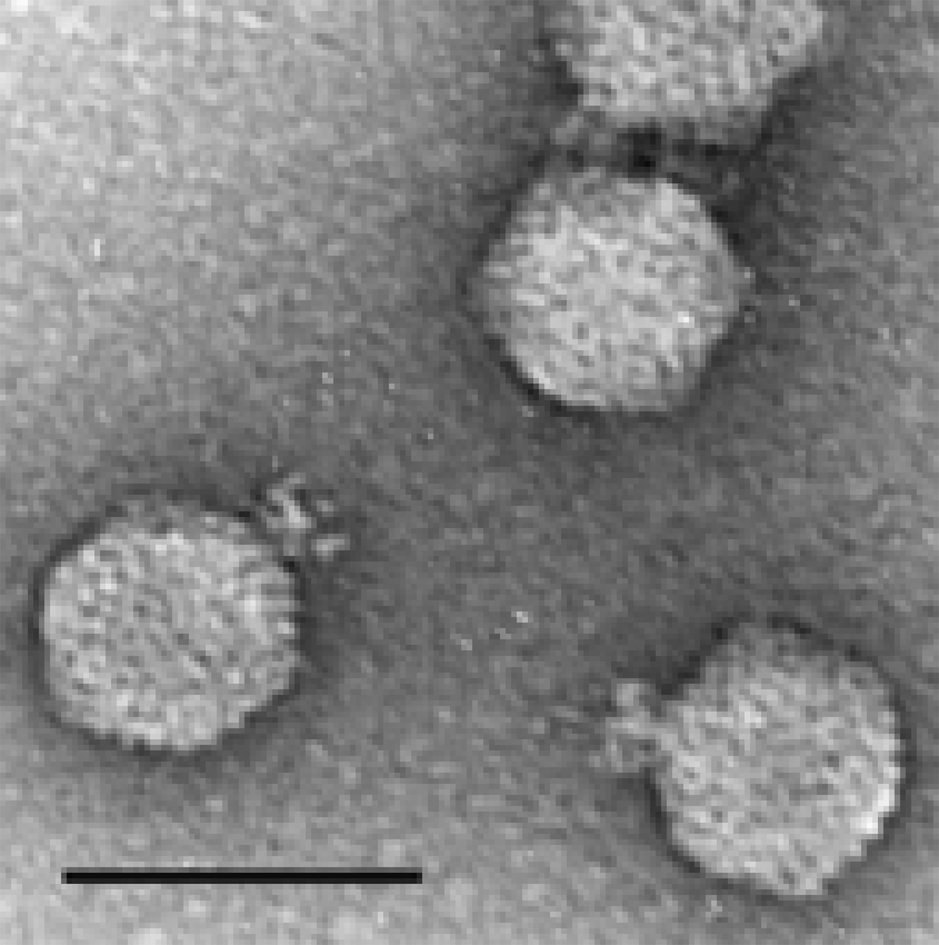
Figure 6 Electron micrograph of phiEco32 stained with phosphotungstate in the presence of bacitracin. The bar represents 100 nm.
(Courtesy Hans-Wolfgang Ackermann.)
Figure 7 (Left) Diagram of particle of Enterobacteria phage T7 (T7) in section. The bar represents 50 nm. (Right) Negative contrast electron micrograph of a particle of phage T7, stained with phosphotungstate. The bar represents 100 nm.
(Modified from Eiserling, F.A. (1979). Bacteriophage structure. In: Comprehensive Virology (H. Fraenkel-Conrat and R.R. Wagner, Eds.), vol. 13, Plenum Press, New York, p. 543; with permission.)
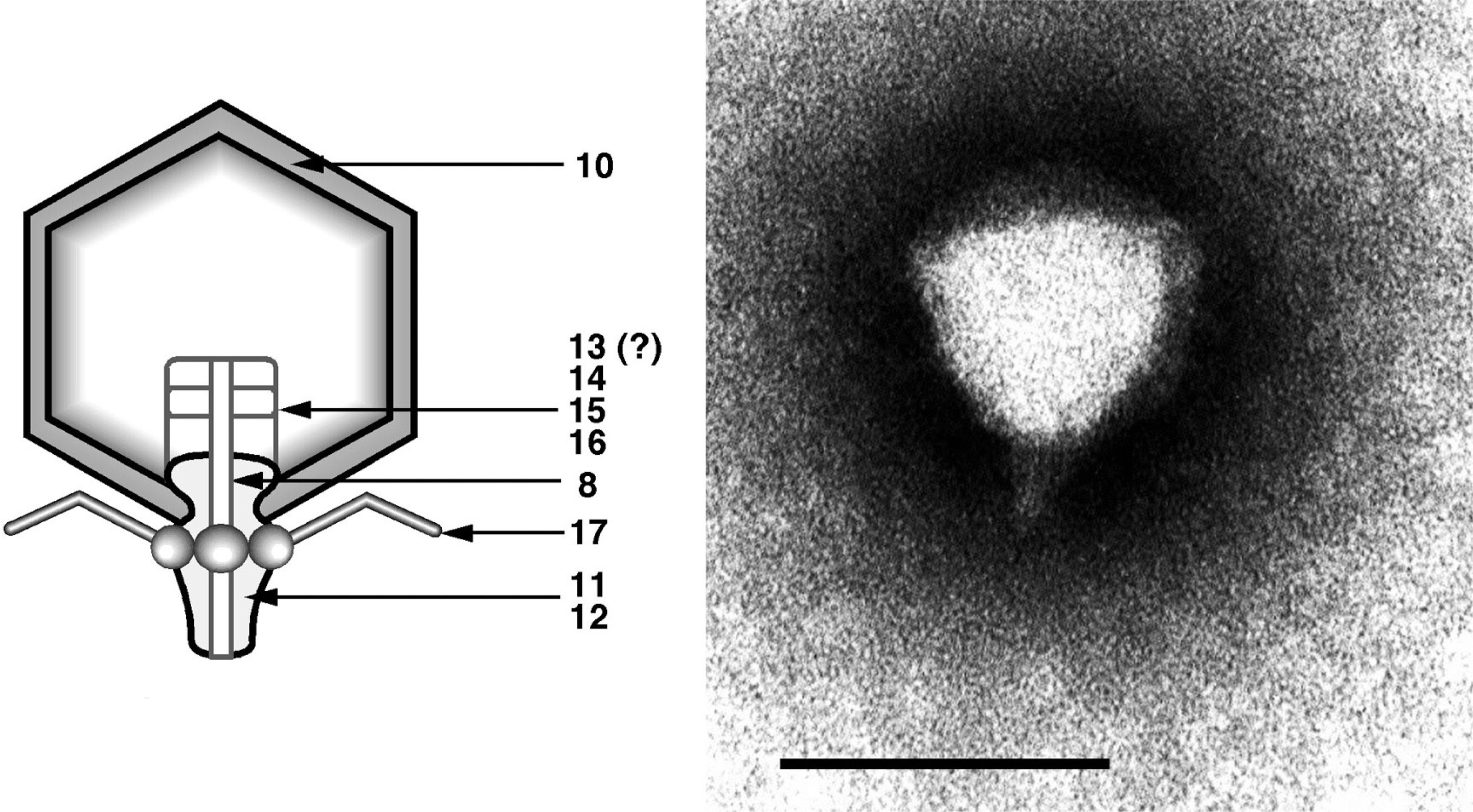
Figure 8 Simplified genetic map of Enterobacteria phage T7 (T7).
(Redrawn after Freifelder, D. (Ed.) (1983). Molecular Biology, Science Books International, Boston, and Van Nostrand Reinhold, New York, p. 630.)
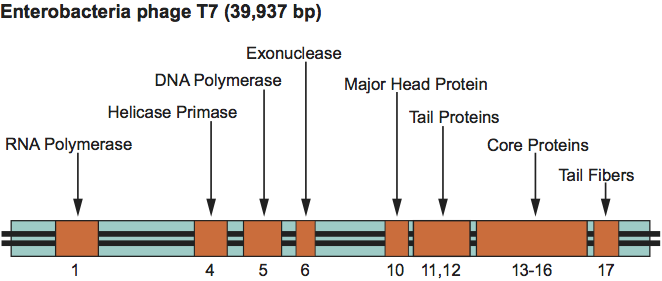
Figure 9 Electron micrograph of the 29-related virus GA-1 stained with phosphotungstate. The bar represents 100 nm.
(Courtesy Hans-Wolfgang Ackermann.)