Family: Plasmaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Plasmavirus
Type species Acholeplasma phage L2
Virion properties
Morphology
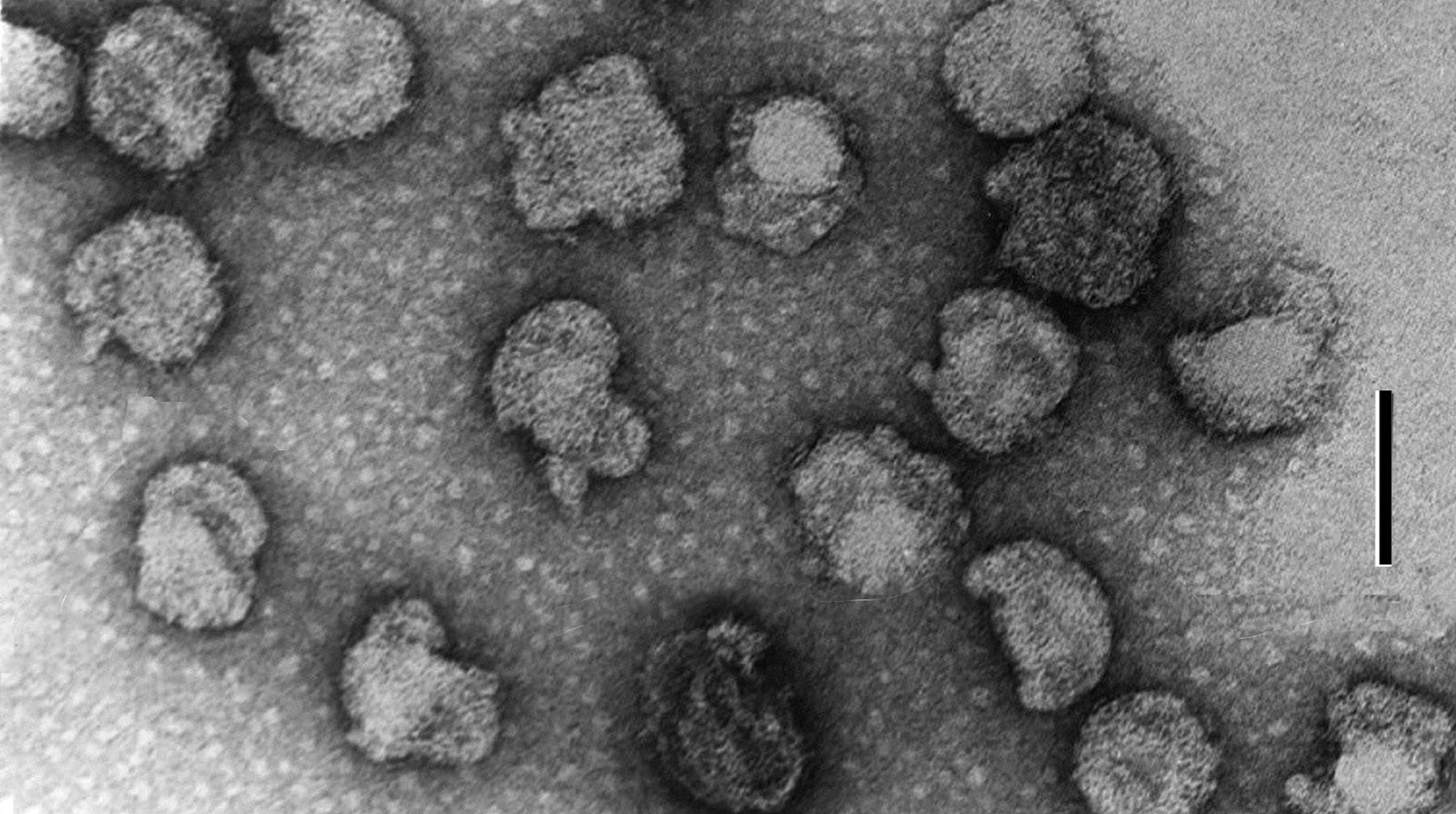
Virions are quasi-spherical, slightly pleomorphic, enveloped and about 80 nm (range 50–125 nm) in diameter (Figure 1). Size varies due to virion heterogeneity: at least three distinct virion forms are produced during infection. Thin-sections show virions with densely stained centers, presumably containing condensed DNA, and particles with lucent centers. The absence of a regular capsid structure suggests the Acholeplasma phage L2 (L2) virion is an asymmetric nucleoprotein condensation bounded by a lipid-protein membrane.
Physicochemical and physical properties
Virions are extremely heat-sensitive, relatively cold-stable and inactivated by nonionic detergents (Brij-58, Triton X-100 and Nonidet P-40), ether and chloroform. Viral infectivity is resistant to DNAse I and phospholipase A, but sensitive to pronase and trypsin treatment. UV-irradiated virions can be reactivated in host cells by excision and SOS DNA repair systems. Virions are relatively resistant to photodynamic inactivation.
Nucleic acid
Virions contain one molecule of infectious, circular, superhelical dsDNA. The phage L2 genome is 11,965 bp, with a G+C value of 32%. All ORFs are encoded in one strand. Several genes are translated from overlapping reading frames.
Proteins
Virions contain at least four major proteins of about 64, 61, 58 and 19 kDa. Several minor protein bands are also observed in virion preparations. DNA sequence analysis indicates 15 ORFs (Table 1).
Table 1 Acholeplasma phage L2 ORFs
| Designation | ORF size (codons) | Comments |
| ORF 1 | 66,643 | – |
| ORF 2 | 9,620 | – |
| ORF 3 | 37,157 | – |
| ORF 4 | 18,224 | – |
| ORF 5 | 34,868 | putative integrase, gene is upstream from attP site |
| ORF 6 | 9,799 | – |
| ORF 7 | 14,047 | – |
| ORF 8 | 7,412 | – |
| ORF 9 | 9,332 | – |
| ORF 10 | 16,143 | – |
| ORF 11 | 25,562 | – |
| ORF 12 | 17,214 | basic protein, putative major virion DNA-binding protein |
| ORF 13 | 81,308 | putative integral membrane protein, has 27 amino acid N-terminal peptidase cleavage signal sequence |
| ORF 13* | 47,699 | translation start site is 295 codons downstream from ORF13 start site and in same reading frame |
| ORF 14 | 26,105 | has 26 aa N-terminal peptidase cleavage signal sequence |
Lipids
Virions and host cell membranes have similar fatty acid compositions. Variation of host cell membrane fatty acid composition leads to virions with corresponding fatty acid composition variations. Data indicate viral membrane lipids are in a bilayer structure.
Carbohydrates
None reported.
Genome organization and replication
L2 infection involves a noncytocidal productive infectious cycle followed by a lysogenic cycle in each infected cell. At least 11 overlapping mRNAs are transcribed from the DNA coding strand, from at least eight promoters. In non-cytocidal infection, progeny phages are released by budding from the host cell membrane, with the host surviving as a lysogen. Lysogeny involves integration of the phage L2 genome into a unique site in the host cell chromosome. The putative phage L2 attP integration site is CATCTTCAT–7nt-CTGAAGATA. Lysogens are resistant to superinfection by homologous virus but not by heterologous virus (apparently due to a repressor), and are inducible by UV-irradiation and mitomycin C.
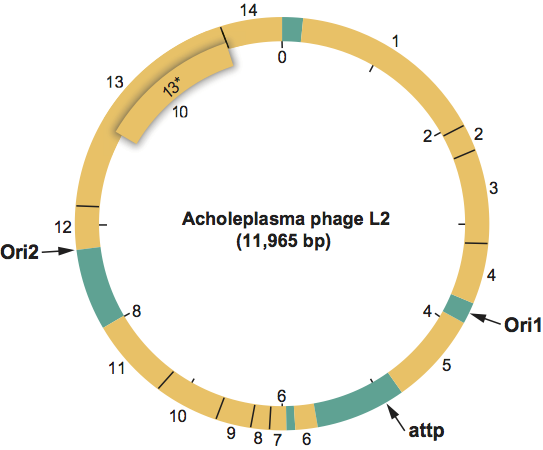
Map of genome organization of Acholeplasma phage L2 (L2) showing ORFs as determined from analysis of the 11,965 bp sequence. The base on the 3’-side of the single BstE II cleavage site is taken as the first base of the DNA sequence. The map also shows locations of the phage L2 integration site (attP) and the two phage L2 DNA replication origin sites (ori1 and ori2).
(From Maniloff et al. (1994); with permission.)
Antigenic properties
None reported.
Biological properties
Host range: the phage L2 infects Acholeplasma laidlawii strains. Other putative plasmaviruses have been reported to infect A. laidlawii (Acholeplasma phages v1, v2, v4, v5 and v7), A. modicum (Acholeplasma phage M1) and A. oculi strains (Acholeplasma phage O1).
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Plasmavirus
| Acholeplasma phage L2 |
|
|
| Acholeplasma phage L2 | [L13696] | (L2) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Plasmavirus but have not been approved as species
| Acholeplasma phage M1 |
| (M1) |
| Acholeplasma phage O1 |
| (O1) |
| Acholeplasma phage v1 |
| (v1) |
| Acholeplasma phage v2 |
| (v2) |
| Acholeplasma phage v4 |
| (v4) |
| Acholeplasma phage v5 |
| (v5) |
| Acholeplasma phage v7 |
| (v7) |
List of unassigned species in the family Plasmaviridae
None reported.
Phylogenetic relationships within the family
No information available.
Similarity with other taxa
None reported.
Derivation of names
Plasma: from the Greek plasma, “shaped product”, referring to the plastic virion shape.
Further reading
Dybvig, K. and Maniloff, J. (1983). Integration and lysogeny by an enveloped mycoplasma virus. J. Gen. Virol., 64, 1781-1785.
Maniloff, J. (1992). Mycoplasma viruses. In: J. Maniloff, R.N. McElhaney, L.R. Finch and J.B. Baseman (Eds.), Mycoplasmas: Molecular Biology and Pathogenesis. American Society for Microbiology, Washington, DC, pp. 41-59.
Maniloff, J., Cadden, S.P. and Putzrath, R.M. (1981). Maturation of an enveloped budding phage: mycoplasma virus L2. In: M.S. DuBow (Ed.), Bacteriophage Assembly, AR Liss Inc., New York, pp. 503-513.
Maniloff, J., Kampo, G.K. and Dascher, C.C. (1994). Sequence analysis of a unique temperate phage: mycoplasma virus L2. Gene, 141, 1-8.
Poddar, S.K., Cadden, S.P., Das, J. and Maniloff, J. (1985). Heterogeneous progeny viruses are produced by a budding enveloped phage. Intervirology, 23, 208-221.
Contributed by
Maniloff, J.
Figures
Figure 1 Negative contrast electron micrograph of Acholeplasma phage L2 (L2) virions. The pleomorphic virion appears as a core (perhaps a nucleoprotein condensation) within a baggy membrane. The bar represents 100 nm.
(From Poddar et al. (1985); with permission.)
Figure 2: Map of genome organization of Acholeplasma phage L2 (L2) showing ORFs as determined from analysis of the 11,965 bp sequence. The base on the 3-side of the single BstE II cleavage site is taken as the first base of the DNA sequence. The map also shows locations of the phage L2 integration site (attP) and the two phage L2 DNA replication origin sites (ori1 and ori2).
(From Maniloff et al. (1994); with permission.)