Family: Phycodnaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Originally, all phycodnavirus virions were assumed to be large icosahedral structures (120–220 nm in diameter) with a multilaminate shell surrounding an electron dense core and lacking an external membrane. However, recent experiments indicate that not all virus structures are identical. Five-fold symmetry averaging 3D reconstruction experiments revealed that one of the vertices in the chlorella virus paramecium bursaria Chlorella virus 1 (PBCV-1) has a cylindrical spike 250 Å long and 50 Å wide (Figure 1A). A possible spike or tail structure is also clearly visible in Emiliania huxleyi virus 86 (EhV-86) during infection (Figure 1B) and tail stubs are often visible in EhV-86 virions. External fibers extend from some of the trisymmetron capsomes of PBCV-1 (probably one per trisymmetron) and may facilitate attachment to the host (Figure 1C). EhV-86 may have an external membrane surrounding the polyhedral capsule (Figure 1D).
Physicochemical and physical properties
The Mr of PBCV-1 is about 1 ×109 and the S20,w is more than 2,000S. Some virions in the genus Chlorovirus are disrupted in CsCl. The infectivity of chloroviruses is not affected by non-ionic detergents but they are inactivated by organic solvents.
Nucleic acid
Virions contain large dsDNA genomes, ranging from 100 to 560 kbp. The G+C content of the viral genomes range from 40 to 52%. The genomic DNA in many of the viruses contains methylated bases, both 5-methylcytosine (m5C) and N6-methyladenine (m6A). The percent of methylated bases in the chloroviruses ranges from no m6A and 0.1% m5C to 37% m6A and 47% m5C.
Proteins
Purified virions contain as many as 100 or more proteins ranging in size from <10 to >200 kDa. The chlorovirus PBCV-1 has three glycoproteins, three myristylated proteins [the major capsid protein, Vp54, is both glycosylated and myristylated] and several phosphoproteins. The Vp54 protein consists of two eight-stranded, antiparallel-beta-barrel, “jelly-roll” domains related by a pseudo six-fold rotation. At least four proteins, including Vp54, are located on the surface of PBCV-1. Proteomic analysis determined that the virion of EhV-86 is composed of at least 28 virus encoded proteins, 23 of which are predicted to be membrane proteins. Besides the major capsid protein, putative function can be assigned to four other components of the virion: two lectin proteins, a thioredoxin and a serine/threonine protein kinase.
Lipids
The chlorovirus PBCV-1 virion contains 5–10% lipid. The lipid is in a bilayer membrane located inside the glycoprotein shell and is required for virus infectivity. The coccolithovirus EhV-86 has an external lipid membrane and may also have an internal membrane (Figure 1D).
Carbohydrates
At least three of the chlorovirus PBCV-1 proteins are glycosylated including the major CP Vp54. The glycan portion of Vp54, which consists of seven neutral sugars, is on the external surface of the virion. Unlike other glycoprotein-containing viruses, PBCV-1 encodes most, if not all, of the machinery required to glycosylate its proteins.
Genome organization and replication
By definition, all phycodnaviruses have large dsDNA genomes. These genomes range in size from 100 kb to over 550 kb with G+C contents ranging from 40 to 52%. Complete genome sequences are available for members in three genera: six chloroviruses, PBCV-1, PBCV-NY2A, PBCV-AR158, PBCV-MT325, PBCV-FR483 and Acanthocystis turfacea chlorella virus 1, two phaeoviruses Ectocarpus siliculosus virus 1 (EsV-1), Feldmannia irregularis virus and the coccolithovirus Emiliania huxleyi virus EhV-86. Partial sequence (approximately 80%) is available for a second coccolithovirus, EhV-163. Additional phycodnavirus genomes are being sequenced, but are not yet publicly available.
There is considerable variation in genome structure among the phycodnaviruses. The PBCV-1 genome is a linear 330 kb, nonpermuted dsDNA molecule with covalently closed hairpin termini (Figure 2, top). The termini consist of 35 nucleotide-long covalently closed hairpin loops that exist in one of two forms; the two forms are complementary when the 35-nucleotide sequences are inverted (flip-flop). Identical 2221-bp inverted repeats are adjacent to each hairpin end. The remainder of the PBCV-1 genome contains primarily single-copy DNA. EsV-1 has a linear dsDNA genome with almost perfect inverted repeats at each end allowing for circularization of the genome (Figure 2, bottom). It is proposed that the inverted repeats anneal with each other to form a cruciform structure that effectively circularizes the genome.
EhV-86 was originally proposed to have a linear genome. However, PCR amplification over the termini revealed a random A/T single nucleotide overhang (50% A, 50% T) suggesting the virus genome has both linear and circular phases. The detection of a DNA ligase and four endonucleases in EhV-86 hints that a linear genome may be packaged in the virions that circularizes during DNA replication.
Repetitive DNA occurs in the PBCV-1, EsV-1 and EhV-86 genomes. Both EsV-1 and PBCV-1 contain about 2 kb inverted repeats adjacent to the terminal ends. In addition to the terminal repeats, tandem repeats are located throughout the EsV-1 genome and comprise approximately 12% of the total genome size. A similar proportion of the Feldmannia sp. virus (FsV) genome also consists of repetitive DNA. EhV-86 has three repeat families (none of which is located at the ends of the genome); one family is postulated to act as an origin of replication (adding credence to the circular mode of replication model), another family is postulated to contain immediate early promoter elements and the last family has a large repetitive proline-rich domain. The repetitive regions in these genomes, while hindering sequencing projects, may play a role in recombination between viruses that allows genetic information to be exchanged with themselves and with their hosts.
Antigenic properties
Four distinct antigenic variants of the chlorovirus PBCV-1 can be isolated that are resistant to polyclonal antibody prepared against wildtype PBCV-1. These variants occur at a frequency of about 1 ×10−6. The antibodies react primarily with the glycan portion of the major CP. Additional variants of these viruses can easily be isolated from natural sources.
Biological properties
The phycodnaviruses, depending on whether they infect freshwater algae or marine algae, are ubiquitous in freshwater or seawater collected throughout the world. Some viruses are host-specific and only infect single isolates or species of algae. For example, chloroviruses only attach to cell walls of certain unicellular, eukaryotic, chlorella-like green algae. Virus attachment is followed by dissolution of the host wall at the point of attachment and entry of the viral DNA and associated proteins into the cell, leaving an empty capsid on the host surface. Beginning about 4 h post infection (p.i.), progeny virions are assembled in the cytoplasm of the host. Infectious virions can be detected inside the cell about 30–40 min prior to virus release; virus release occurs by cell lysis. Coccolithoviruses, prymnesioviruses and raphidoviruses have wider host ranges, where individual viruses can infect a range of host isolates within specific algal species; however they do not cross the species barrier.
The phaeoviruses infect the wall-less spore or gamete stage of filamentous brown algae, followed by fusion of adjacent host and particle surfaces. Empty particles remain on the cell surface following the release of core contents. An eclipse period of approximately 3 h follows the attachment stage. The virus growth cycle is complete after approximately 14 h. During the replication cycle, particles appear in the cytoplasm and are associated with the production of cytoplasmic fibrils (ca. 5–8 nm in diameter) and clusters of membrane-bound vesicles that are absent in healthy cells. Particles are released into the medium via localized ruptures in the cell membrane; ruptures often appear at several locations on the same cell.
Coccolithoviruses attach to exposed membranes of their host and the viruses enter into the host intact via either an endocytotic or an envelope fusion mechanism, after which they rapidly disassemble (Figure 3).
Less is known about the replication of prymnesioviruses and raphidoviruses. Virus formation is observed in the cytoplasm and the nucleus remains intact and separate from the viroplasm that consists of a fibrillar matrix. Ultimately, viral production results in the disruption of organelles, lysis of the cell and release of the virus particles.
The hosts for some of the chloroviruses and coccolithoviruses can easily be grown in the laboratory and the viruses can be plaque-assayed. The hosts for some of the other viruses are either cultured axenically (e.g., prymnesiovirus hosts, P. globosa) or non-axenically in uni-alga cultures (e.g. hosts for the prasinoviruses and raphidoviruses). The brown algal viruses, which only appear in mature gametangia or sporangia cells of their hosts, can also be grown in the laboratory.
The chloroviruses, coccolithoviruses, prasinoviruses, prymnesioviruses and raphidoviruses are transmitted horizontally. The phaeoviruses are transmitted both horizontally and vertically.
Genus Chlorovirus
Type species Paramecium bursaria Chlorella virus 1
Distinguishing features
The chloroviruses, which are ubiquitous in freshwater throughout the world, infect certain unicellular, eukaryotic, exsymbiotic chlorella-like green algae. The viruses have linear, non-permuted dsDNA genomes with cross-linked hairpin ends. The DNA termini contain identical inverted 1–2.2 kb repeats. The remainder of the genome is primarily single copy DNA. Viruses in this group can be distinguished from each other by DNA restriction digests, the levels of methylated bases in their genomes, serology and host specificity.
Originally the chloroviruses were believed to be simple polyhedral structures (ca. 190 nm in diameter) with a multilaminate shell surrounding an internal bilayered membrane. However, five-fold symmetry averaging 3D reconstruction experiments of PBCV-1 revealed that one of the vertices in the chlorella viruses has a cylindrical spike 250 Å long and 50 Å wide (Figure 1A). The spike is too narrow for DNA to pass through it. External fibers extend from some of the trisymmetron capsomes (probably one per trisymmetron) and probably facilitate attachment to the host (Figure 1B). PBCV-1 initiates infection by attaching rapidly and specifically to the host cell wall, probably by the fibers mentioned above. Following host cell wall degradation by virus-packaged enzyme(s), the PBCV-1 internal membrane presumably fuses with the host membrane, facilitating entry of the viral DNA and virion-associated proteins into the cell, leaving an empty capsid attached to the surface. PBCV-1 lacks a recognizable RNA polymerase gene, and so circumstantial evidence suggests that the virus DNA and DNA-associated proteins quickly move to the nucleus, where early transcription begins within a few minutes. PBCV-1 DNA replication begins 60–90 min p.i. and is followed by transcription of late genes. Approximately 2–3 h p.i. assembly of virus capsids begin in localized regions in the cytoplasm, which become prominent 3–4 h p.i. Five to six hours p.i. the cytoplasm fills with infectious progeny virus particles and localized lysis of the host cell releases progeny at 6–8 h p.i.
The 330 kb PBCV-1 genome, along with five other chloroviruses, have been sequenced. The viruses encode as many as 800 ORFs, 40 codons or longer, of which about 50% are predicted to encode proteins. About 40% of these putative protein-encoding ORFs match proteins in the databases. Two of the PBCV-1 genes are interrupted by introns: a transcription factor TFIIS-like gene has a self-splicing type I intron and the DNA polymerase gene has a spliceosomal processed type of intron. The PBCV-1 genome also has 11 tRNA genes, one of which is predicted to contain a small intron. In addition, one of the chloroviruses, NY-2A, contains two inteins. The chloroviruses encode many interesting and unusual proteins including DNA restriction endonucleases, polyamine biosynthetic enzymes, sugar metabolizing enzymes, and ion channel and transporting proteins.
Species demarcation criteria in the genus
Three groups of viruses are delineated based on host specificity:
Group 1. Paramecium bursaria Chlorella NC64A viruses (NC64A viruses)
Group 2. Paramecium bursaria Chlorella Pbi viruses (Pbi viruses)
Group 3. Hydra viridis Chlorella viruses (HVC viruses)
Chlorella strains NC64A, ATCC 30562 and N1A (originally symbionts of the protozoan P. bursaria), collected in the United States, are the only known host for NC64A viruses. Chlorella strain Pbi (originally a symbiont of a European strain of P. bursaria) collected in Germany, is the only known host for Pbi viruses. Pbi viruses do not infect Chlorella strains NC64A, ATCC 30562 and N1A. Chlorella strain Florida (originally a symbiont of Hydra viridis) is the only known host for Hydra viridis Chlorella virus (HVCV). NC64A viruses are grouped into 16 species based on plaque size, serological reactivity, resistance of the genome to restriction endonucleases, virus encoded restriction endonucleases and nature and content of methylated bases.
List of species in the genus Chlorovirus
| Group 1: Paramecium bursaria Chlorella NC64A virus group |
|
|
| Paramecium bursaria Chlorella virus 1 |
|
|
| Paramecium bursaria Chlorella virus 1 | [U42580] | (PBCV-1) |
| Paramecium bursaria Chlorella virus AL1A |
|
|
| Paramecium bursaria Chlorella virus AL1A |
| (PBCV-AL1A) |
| Paramecium bursaria Chlorella virus AL2A |
|
|
| Paramecium bursaria Chlorella virus AL2A |
| (PBCV-AL2A) |
|
|
|
|
| Paramecium bursaria Chlorella virus AR158 | [NC_009899] | (PBCV-AR158) |
| Paramecium bursaria Chlorella virus BJ2C |
|
|
| Paramecium bursaria Chlorella virus BJ2C |
| (PBCV-BJ2C) |
| Paramecium bursaria Chlorella virus CA4A |
|
|
| Paramecium bursaria Chlorella virus CA4A |
| (PBCV-CA4A) |
| Paramecium bursaria Chlorella virus CA4B |
|
|
| Paramecium bursaria Chlorella virus CA4B |
| (PBCV-CA4B) |
| Paramecium bursaria Chlorella virus IL3A |
|
|
| Paramecium bursaria Chlorella virus IL3A |
| (PBCV-IL3A) |
| Paramecium bursaria Chlorella virus NC1A |
|
|
| Paramecium bursaria Chlorella virus NC1A |
| (PBCV-NC1A) |
| Paramecium bursaria Chlorella virus NE8A |
|
|
| Paramecium bursaria Chlorella virus NE8A |
| (PBCV-NE8A) |
| Paramecium bursaria Chlorella virus NY2A |
|
|
| Paramecium bursaria Chlorella virus NY2A | [DQ491002] | (PBCV-NY2A) |
| Paramecium bursaria Chlorella virus NYs1 |
|
|
| Paramecium bursaria Chlorella virus NYs1 |
| (PBCV-NYs1) |
| Paramecium bursaria Chlorella virus SC1A |
|
|
| Paramecium bursaria Chlorella virus SC1A |
| (PBCV-SC1A) |
| Paramecium bursaria Chlorella virus XY6E |
|
|
| Paramecium bursaria Chlorella virus XY6E |
| (PBCV-XY6E) |
| Paramecium bursaria Chlorella virus XZ3A |
|
|
| Paramecium bursaria Chlorella virus XZ3A |
| (PBCV-XZ3A) |
| Paramecium bursaria Chlorella virus XZ4A |
|
|
| Paramecium bursaria Chlorella virus XZ4A |
| (PBCV-XZ4A) |
| Paramecium bursaria Chlorella virus XZ4C |
|
|
| Paramecium bursaria Chlorella virus XZ4C |
| (PBCV-XZ4C) |
| Group 2: Paramecium bursaria Chlorella Pbi virus group |
|
|
| Paramecium bursaria Chlorella virus A1 |
|
|
| Paramecium bursaria Chlorella virus A1 |
| (PBCV-A1) |
|
|
|
|
| Paramecium bursaria Chlorella virus FR483 | [NC_008603] | (PBCV-FR483) |
|
|
|
|
| Paramecium bursaria Chlorella virus MT325 | [DQ491001] | (PBCV-MT325) |
| Group 3: Hydra viridis Chlorella virus group |
|
|
| Hydra viridis Chlorella virus 1 |
|
|
| Hydra viridis Chlorella virus 1 |
| (HVCV-1) |
| Group 4: Acanthocystis turfacea chlorella virus group |
|
|
| Acanthocystis turfacea chlorella virus 1 |
|
|
| Acanthocystis turfacea chlorella virus 1 | [EF101928] | (ATCV-1) |
Species names are in italic script; names of strains and isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Chlorovirus but have not been approved as species
| Paramecium bursaria Chlorella virus AL2C |
| (PBCV-AL2C) |
| Paramecium bursaria Chlorella virus CA1A |
| (PBCV-CA1A) |
| Paramecium bursaria Chlorella virus CA1D |
| (PBCV-CA1D) |
| Paramecium bursaria Chlorella virus CA2A |
| (PBCV-CA2A) |
| Paramecium bursaria Chlorella virus IL2A |
| (PBCV-IL2A) |
| Paramecium bursaria Chlorella virus IL2B |
| (PBCV-IL2B) |
| Paramecium bursaria Chlorella virus IL3D |
| (PBCV-IL3D) |
| Paramecium bursaria Chlorella virus IL5-2s1 |
| (PBCV-IL5-2s1) |
| Paramecium bursaria Chlorella virus MA1D |
| (PBCV-MA1D) |
| Paramecium bursaria Chlorella virus MA1E |
| (PBCV-MA1E) |
| Paramecium bursaria Chlorella virus NC1B |
| (PBCV-NC1B) |
| Paramecium bursaria Chlorella virus NC1C |
| (PBCV-NC1C) |
| Paramecium bursaria Chlorella virus NC1D |
| (PBCV-NC1D) |
| Paramecium bursaria Chlorella virus NE8D |
| (PBCV-NE8D) |
| Paramecium bursaria Chlorella virus NY2B |
| (PBCV-NY2B) |
| Paramecium bursaria Chlorella virus NY2C |
| (PBCV-NY2C) |
| Paramecium bursaria Chlorella virus NY2F |
| (PBCV-NY2F) |
| Paramecium bursaria Chlorella virus NYb1 |
| (PBCV-NYb1) |
| Paramecium bursaria Chlorella virus SC1B |
| (PBCV-SC1B) |
| Paramecium bursaria Chlorella virus SH6A |
| (PBCV-SH6A) |
| Paramecium bursaria Chlorella virus XZ5C |
| (PBCV-XZ5C) |
| Paramecium bursaria Chlorella virus CVBII |
| (PBCV-CVBII) |
| Paramecium bursaria Chlorella virus CVK2 |
| (PBCV-CVK2) |
| Paramecium bursaria Chlorella virus CVU1 |
| (PBCV-CVU1) |
Genus Coccolithovirus
Type species Emiliania huxleyi virus 86
Distinguishing features
Viruses assigned to this genus all have large dsDNA genomes (ca. 410–415 kbp) and probably have a common genome structure. The particles have icosohedral symmetry, enveloped by a lipid membrane, are tailless and range from 150 to 200 nm in diameter. The latent period of these viruses is 3–4 h and the burst size is 400–1000 viruses per lysed host cell (mean 620). They infect different isolates of the globally important marine coccolithophorid Emiliania huxleyi, a marine alga which has a world-wide distribution and is known for forming vast coastal and mid-oceanic blooms which are easily observed by satellite imagery. The viruses described here were isolated from E. huxleyi blooms off the coast of Plymouth, UK, in July 1999 and July/August 2001, from an E. huxleyi bloom induced during a mesocosm experiment in a fjord near Bergen, Norway, during June 2000 and from E. huxleyi blooms off the coast of Bergen, Norway, in June 1999 and June 2000. The viruses are relatively easy to isolate and susceptible host strains usually lyse 2–7 days after the addition of filtered seawater. Clonal isolates can be obtained by plaque assay.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Coccolithovirus
| Emiliania huxleyi virus 86 |
|
|
| Emiliania huxleyi virus 86 | [AJ890364] | (EhV-86) |
Species names are in italic script; names of strains and isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Coccolithovirus but have not been approved as species
| Emiliania huxleyi virus 84 |
| (EhV-84) |
| Emiliania huxleyi virus 88 |
| (EhV-88) |
| Emiliania huxleyi virus 163 |
| (EhV-163) |
| Emiliania huxleyi virus 201 |
| (EhV-201) |
| Emiliania huxleyi virus 202 |
| (EhV-202) |
| Emiliania huxleyi virus 203 |
| (EhV-203) |
| Emiliania huxleyi virus 205 |
| (EhV-205) |
| Emiliania huxleyi virus 207 |
| (EhV-207) |
| Emiliania huxleyi virus 208 |
| (EhV-208) |
| Emiliania huxleyi virus 99B1 | [FN429076] | (EhV-99B1) |
| Emiliania huxleyi virus 2KB1 |
| (EhV-2KB1) |
| Emiliania huxleyi virus 2KB2 |
| (EhV-2KB2) |
| Emiliania huxleyi virus Ø28 |
| (EhV- Ø28) |
| Emiliania huxleyi virus Ø29 |
| (EhV- Ø29) |
| Emiliania huxleyi virus Ø30 |
| (EhV- Ø30) |
| Emiliania huxleyi virus Ø42 |
| (EhV- Ø42) |
| Emiliania huxleyi virus Ø43 |
| (EhV- Ø43) |
Genus Prasinovirus
Type species Micromonas pusilla virus SP1
Distinguishing features
Viruses assigned to this genus infect marine prasinophytes from the three genera Bathycoccus, Micromonas and Ostreococcus, the latter being the world’s smallest free living eukaryote. With a small host size (less than 1 μm in diameter for Ostreococcus) and a virus capsid size of around 120 nm, it is estimated there is physically room for no more than 100 virions at any one time. This is reflected in experimental data that suggest a typical burst size of 6–15 viruses per cell. Following viral adsorption, genome replication occurs from 2 hours post infection (p.i.), virions assemble in the cytoplasm from 6 h p.i. until 20 h p.i., after which cellular lysis occurs. The host cell nucleus, mitochondria and chloroplast remain intact through this period. The prasinoviruses have genomes in the size range of 184–198 kbp and are morphologically similar to other phycodnaviruses with icosahedral capsids of 100–130 nm. The viruses are ubiquitous in seawater throughout the world, and have been isolated from the Pacific and Atlantic Oceans, the Gulf of Mexico and the Mediterranean Sea.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Prasinovirus
| Micromonas pusilla virus SP1 |
|
|
| Micromonas pusilla virus SP1 |
| (MpV-SP1) |
Species names are in italic script; names of strains and isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Prasinovirus but have not been approved as species
| Bathycoccus prasinos virus 1 | [HM004432] | (BpV-1) |
| Bathycoccus prasinos virus 1 | [HM004430] | (BpV-2) |
| Micromonas pusilla virus 1 | [HM004429] | (MpV-1) |
| Micromonas pusilla virus GM1 |
| (MpV-GM1) |
| Micromonas pusilla virus PB6 |
| (MpV-PB6) |
| Micromonas pusilla virus PB7 |
| (MpV-PB7) |
| Micromonas pusilla virus PB8 |
| (MpV-PB8) |
| Micromonas pusilla virus PL1 |
| (MpV-PL1) |
| Micromonas pusilla virus SG1 |
| (MpV-SG1) |
| Micromonas pusilla virus SP2 |
| (MpV-SP2) |
| Ostreococcus luminaris virus 1 | [HM004431] | (OlV-1) |
| Ostreococcus tauri virus 1 | [FN386611] | (OtV-1) |
| Ostreococcus tauri virus 2 | [FN600414] | (OtV-2) |
| Ostreococcus tauri virus 5 | [EU304328] | (OtV-5) |
Genus Prymnesiovirus
Type species Chrysochromulina brevifilum virus PW1
Distinguishing features
Viruses assigned to this genus infect hosts belonging to algal class Haptophyceae (also referred to as the Prymnesiophyceae). Although viruses in this genus all have dsDNA genomes, there is a wide variety of diameters (100–170 nm) and genome sizes (120–485 kbp). Estimated burst sizes range from 320 to 600 viruses per infected cell. Viruses that infect members of the same marine algae, Chysochromulina brevifilum and C. strobilus, were isolated from USA (Texas) coastal waters in three locations (Gulf of Mexico, Aransas Pass and Laguna Madre). Viruses that infect Phaeocystis globosa were isolated from natural seawater off the coast of the Netherlands during April to July 2000 and 2001, the North Sea in April 2002, from a P. globosa bloom induced during a mesocosm experiment during October 2000 and from a P. globosa bloom in the English Channel off the coast of Plymouth, UK, in April 2001. These viruses lysed susceptible host strains after 2–7 days after the addition of filtered seawater. Once isolated, susceptible host strains typically lyse within 1–2 days.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Prymnesiovirus
| Chysochromulina brevifilum virus PW1 |
|
|
| Chysochromulina brevifilum virus PW1 |
| (CbV-PW1) |
Species names are in italic script; names of strains and isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Prymnesiovirus but have not been approved as species
| Chysochromulina brevifilum virus PW3 |
| (CbV-PW3) |
| Phaeocystis globosa virus 1 (Texel) |
| (PgV-01T) |
| Phaeocystis globosa virus 2 (Texel) |
| (PgV-02T) |
| Phaeocystis globosa virus 3 (Texel) |
| (PgV-03T) |
| Phaeocystis globosa virus 4 (Texel) |
| (PgV-04T) |
| Phaeocystis globosa virus 5 (Texel) |
| (PgV-05T) |
| Phaeocystis globosa virus 6 (Texel) |
| (PgV-06T) |
| Phaeocystis globosa virus 7 (Texel) |
| (PgV-07T) |
| Phaeocystis globosa virus 9 (Texel) |
| (PgV-09T) |
| Phaeocystis globosa virus 10 (Texel) |
| (PgV-10T) |
| Phaeocystis globosa virus 11 (Texel) |
| (PgV-11T) |
| Phaeocystis globosa virus 12 (Texel) |
| (PgV-12T) |
| Phaeocystis globosa virus 13 (Texel) |
| (PgV-13T) |
| Phaeocystis globosa virus 14 (Texel) |
| (PgV-14T) |
| Phaeocystis globosa virus 15 (Texel) |
| (PgV-15T) |
| Phaeocystis globosa virus 16 (Texel) |
| (PgV-16T) |
| Phaeocystis globosa virus 17 (Texel) |
| (PgV-17T) |
| Phaeocystis globosa virus 18 (Texel) |
| (PgV-18T) |
| Phaeocystis globosa virus 102 (Plymouth) |
| (PgV-102P) |
Genus Phaeovirus
Type species Ectocarpus siliculosus virus 1
Distinguishing features
Ectocarpus siliculosus virus 1 isolates infect the free-swimming, zoospore or gamete stages of filamentous brown algal hosts. The virus genome is integrated into the host genome and is inherited in a Mendelian manner. Virus particles are only formed in prospective gametangia or sporangia cells of the host. Phaeoviruses share icosahedral morphologies with internal lipid membranes and large, complex, double stranded DNA genomes. The replication strategy for these virus genomes is yet to be resolved but their genomes occur in both linear and circular forms.
Species demarcation criteria in the genus
Nine species of viruses are delineated based in part on host specificity. Field isolates of at least seven genera of the Phaeophycae contain 120–150 nm diameter polyhedral virus-like particles. The particles contain dsDNA genomes that vary in size from 150 to 350 kb, although the major CP gene and DNA polymerase sequence data indicate that they are closely related. Virus expression is variable; particles are rarely observed in vegetative cells but are common in unilocular sporangia (Feldmannia species virus, FsV) or both unilocular and plurilocular sporangia and gametangia (EsV). Some of the viruses have a narrow host range (FsV), whereas others such as Ectocarpus fasciculatus virus (EfV) and EsV infect members of more than one genus.
List of species in the genus Phaeovirus
| Ectocarpus fasciculatus virus a |
|
|
| Ectocarpus fasciculatus virus 1 |
| (EfV-1) |
| Ectocarpus siliculosus virus 1 |
|
|
| Ectocarpus siliculosus virus 1 | [AF204951] | (EsV-1) |
| Ectocarpus siliculosus virus 1-like (pro-virus) |
| (EsV-1) |
| Ectocarpus siliculosus virus a |
|
|
| Ectocarpus siliculosus virus a |
| (EsV-1a) |
| Feldmannia irregularis virus a |
|
|
| Feldmannia irregularis virus 1 |
| (FirrV-1) |
| Feldmannia species virus |
|
|
| Feldmannia species virus 158 | [EU916176] | (FsV-158) |
| Feldmannia species virus 178 |
| (FsV-178) |
| Hincksia hinckiae virus a |
|
|
| Hincksia hinckiae virus 1 |
| (HhV-1) |
| Myriotrichia clavaeformis virus a |
|
|
| Myriotrichia clavaeformis virus 1 |
| (McV-1) |
| Pilayella littoralis virus 1 |
|
|
| Pilayella littoralis 1 |
| (PlV-1) |
Species names are in italic script; names of strains and isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Phaeovirus but have not been approved as species
None reported.
Genus Raphidovirus
Type species Heterosigma akashiwo virus 01
Distinguishing features
Viruses assigned to this genus infect the harmful bloom causing raphidophyte, Heterosigma akashiwo (Raphidophyceae), a marine alga that has a world-wide distribution. Members of the type species Heterosigma akashiwo virus 01 have a large dsDNA genome (ca. 294 kbp) and virions with icosahedral symmetry that are 202±6 nm in diameter. The viruses were isolated from H. akashiwo blooms near the western coast of Japan. The latent period and the burst size of HaV01 is 30–33 h and about 770 at 20 °C, respectively. Although these viruses rapidly degrade to lose infectivity even when kept at 4 °C in the dark, they can be easily cryopreserved. Their infection is considered to be a significant factor influencing the dynamics and termination of H. akashiwo blooms. HaV infection has great impacts on H. akashiwo populations regarding both the fluctuation of the algal biomass (quantity) and the changes in the clonal composition (quality); the latter is most likely due to the variety in interspecies host specificity of HaV clones. Partial sequencing of the HaV01 genome revealed that several genes resembled other protist-infecting nucleocytoplasmic large DNA viruses. Interestingly, the genome included a 232-amino-acid intein (protein intron) in its DNA polymerase gene.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Raphidovirus
| Heterosigma akashiwo virus 01 |
|
|
| Heterosigma akashiwo virus 01 |
| (HaV-01) |
Species names are in italic script; names of strains and isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Raphidovirus but have not been approved as species
None reported.
List of unassigned species in the family Phycodnaviridae
None reported.
List of other related viruses which may be members of the family Phycodnaviridae but have not been approved as species
The following viruses are considered potential candidates for assignment to the family Phycodnaviridae. However, this cannot be confirmed without further characterization.
Aureococcus anophagefference virus (AaV) (Brown tide virus): This virus was isolated from Great South Bay New York, USA, in 1992. Viruses are 140–160 nm in diameter and cultures of Aureococcus anophagefference usually lyse within 24–48 h.
Chrysochromulina ericina virus 01B (CeV-01B): This virus was isolated from water collected off the coast of Bergen, Norway, in June 1998. Viruses have particle diameters of 160 nm and a genome size of 510 kbp. The lytic cycle is 14–19 h and the burst size is about 1800–4100 viruses per host cell. Although included in the phylogenetic analysis for the Phycodnaviridae (Figure 4), the taxonomic position of CeV taxonomic is under debate.
Heterocapsa circularisquama viruses (HcV-01 to HcV-10): These viruses have been isolated from H. circularisquama blooms in the coastal waters of Japan. They have a large dsDNA genome (ca. 350 kbp), icosahedral symmetry and are 180–210 nm in diameter (197±8 nm). The latent period and the burst size is 40–56 h and about 1800–2400 infectious units at 20–25 °C, respectively. Although these viruses rapidly degrade to lose infectivity even when kept at 4 °C in the dark, they can be cryopreserved. Classification of HcV is currently under debate since phylogenetic analyses of the HcV DNA polymerase indicates a high similarity with African swine fever virus (genus Asfarvirus, family Asfarviridae).
Phaeocystis pouchetii virus 01 (PpV-01): This virus was isolated from water collected at the end of a P. pouchetii bloom off the coast of Bergen, Norway, in May 1995. Viruses have particle diameters of 130–160 nm and a genome size of 485 kbp. The latent period is 12–18 h, complete lysis of cultures is observed after 48 h and the burst size is about 350–600 viruses per host cell. The taxonomic position of PgV is under debate since it falls in the same clade as the family Mimiviridae.
Pyramimonas orientalis virus 01B (PoV-01B): This virus was isolated from water collected off the coast of Bergen, Norway, in June 1998. Viruses have particle sizes of 220×180 nm and a genome size of 560 kbp. The lytic cycle is 14–19 h and the burst size is about 800–1000 viruses per host cell. Although included in the phylogenetic analysis for the Phycodnaviridae (Figure 4), the taxonomic position of PoV is under debate.
| Aureococcus anophagefference virus (Brown tide virus) |
| (AaV) |
| Chrysochromulina ericina virus 01B |
| (CeV-01B) |
| Heterocapsa circularisquama virus 01 |
| (HcV-01) |
| Heterocapsa circularisquama virus 02 |
| (HcV-02) |
| Heterocapsa circularisquama virus 03 |
| (HcV-03) |
| Heterocapsa circularisquama virus 04 |
| (HcV-04) |
| Heterocapsa circularisquama virus 05 |
| (HcV-05) |
| Heterocapsa circularisquama virus 06 |
| (HcV-06) |
| Heterocapsa circularisquama virus 07 |
| (HcV-07) |
| Heterocapsa circularisquama virus 08 |
| (HcV-08) |
| Heterocapsa circularisquama virus 09 |
| (HcV-09) |
| Heterocapsa circularisquama virus 10 |
| (HcV-10) |
| Phaeocystis pouchetii virus 01 |
| (PpV-01) |
| Pyramimonas orientalis virus 01B |
| (PoV-01B) |
Phylogenetic relationships within the family
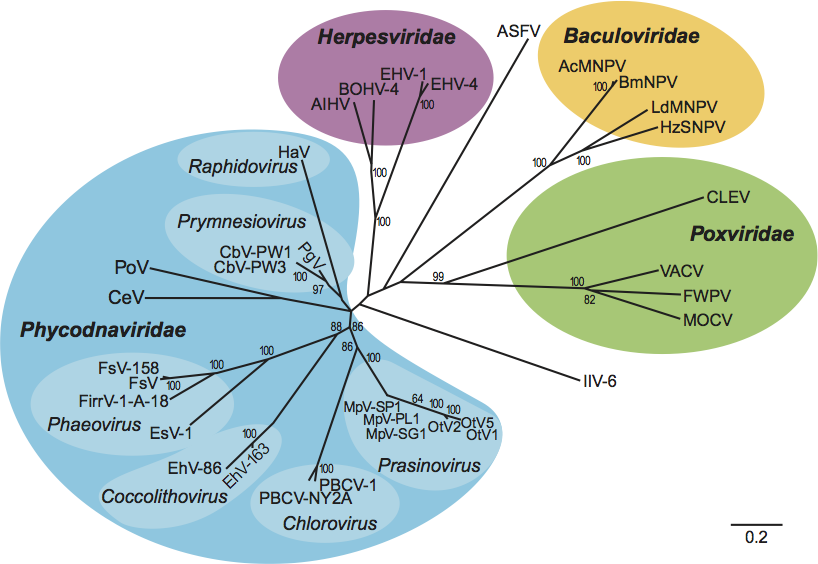
At the time of writing, the phylogeny of the Phycodnaviridae is in question, owing to the wide variation in DNA sequence that is observed between the six clades of the family Phycodnaviridae (ringed in Figure 4; DNA sequence identities range from 29% to 98%). It is possible that the Phycodnaviridae may need to be split into separate families as more sequence data become available (e.g. see discussion by Moreau et al., 2010).
Similarity with other taxa
See comments above about phylogenetic relationships. In addition, many large polyhedral virus-like particles have been observed in electron micrographs of eukaryotic algae. However, for the most part these particles have not been characterized.
Derivation of names
Chloro: from Greek chloro, “green”.
Cocco: derived from Greek kokkis, “grain” or “berry” (referring to their shape).
dna: for deoxyribonucleic acid.
Lith: from Greek Lithos, “stone”.
Phaeo: from Greek phaeo, “brown”.
Phyco: from Greek phycos, “plant”.
Prasino: from Latin prasino, “green”.
Prymnesio: from Greek prymne, “stern of a ship”.
Raphido: from Greek raphido, “spine”.
Further reading
Chen, F., Suttle, C.A., 1996. Evolutionary relationships among large double-stranded DNA viruses that infect microalgae and other organisms as inferred from DNA polymerase genes. Virology. 219, 170–178.
Derelle, E., Ferraz, C., Escande, M.-L., Eychenia, S., Cooke, R., Piganeau, G.L., Desdevises, Y., Bellec, L., Moreau, H., Grimsley, N., 2008. Life-cycle and genome of OtV5, a large DNA virus of the pelagic marine unicellular green alga Ostreococcus tauri. PLoS ONE. 3, e2250.
Dunigan, D.D., Fitzgerald, L.A., Van Etten, J.L., 2006. Phycodnaviruses: a peek at genetic diversity. Virus Res. 117, 119–132.
Moreau, H., Piganeau, G., Desdevises, Y., Cooke, R., Derelle, E., Grimsley, N., 2010. Marine prasinovirus genomes show low evolutionary divergence and acquisition of protein metabolism genes by horizontal gene transfer. J. Virol. 84, 12555–12563.
Sandaa, R.A., Heldal, M., Castberg, T., Thyrhaug, R., Bratbak, G., 2001. Isolation and characterization of two viruses with large genome size infecting Chrysochromulina ericina (Prymnesiophyceae) and Pyramimonas orientalis (Prasinophyceae). Virology. 290, 272–280.
Schroeder, D.C., Oke, J., Malin, G., Wilson, W.H., 2002. Coccolithovirus (Phycodnaviridae): Characterisation of a new large dsDNA algal virus that infects Emiliania huxleyi. Arch. Virol. 147, 1685–1698.
Schroeder, D.C., Park, Y., Yoon, H-M, Lee, Y.S., Kang, S.W., Meints, R.H., Ivey, R.G., Choi, T-J., 2009. Genomic analysis of the smallest giant virus - Feldmannia sp. virus 158. Virology. 384, 223–232.
Tomaru, Y., Shirai, Y., Nagasaki, K., 2008. Ecology, physiology and genetics of a phycodnavirus infecting the noxious bloom-forming raphidophyte Heterosigma akashiwo. Fisheries Sci. 74, 701–711.
Van Etten, J.L., 2003. Unusual life style of giant chlorella viruses. Annu. Rev. Genet. 37, 153–195.
Wilson, W.H., Van Etten, J.L., Allen, M.J., 2009. The Phycodnaviridae: the story of how tiny giants rule the world. In: Van Etten, J., Lesser Known Large dsDNA Viruses. Springer, Berlin, 1–42.
Contributed by
Wilson, W.H., Van Etten, J.L., Schroeder, D.C., Nagasaki, K., Brussaard, C., Bratbak, G. and Suttle, C.
Figures
Figure 1 (A) Fivefold averaged cryo-electron micrographs of Paramecium bursaria chlorella virus PBCV-1 reveal a long, thin, cylindrical spike structure at one vertex and protrusions (fibers) extending from one unique capsomer per trisymmetron (from Cherrier et al. (2009). Proc. Natl Acad. Sci., U S A, 106, 11085-89; with permission). (B) Putative tail structure (arrowed) can be observed in Emiliania huxleyi virus EhV-86 in the cytoplasm of infected Emiliania huxleyi before release of progeny virions (approx. 3 h p.i.) (adapted from Mackinder et al. (2010). J. Gen. Virol., 90, 2306-2316; with permission). (C) PBCV-1 attached to the cell wall of its host as viewed by the quick-freeze, deep-etch procedure. Note: fibers attach the virus to the wall (photo courtesy of John Heuser). (D) EhV-86 virion showing the putative internal lipid membrane (arrowed)
(photo courtesy of Willie Wilson).
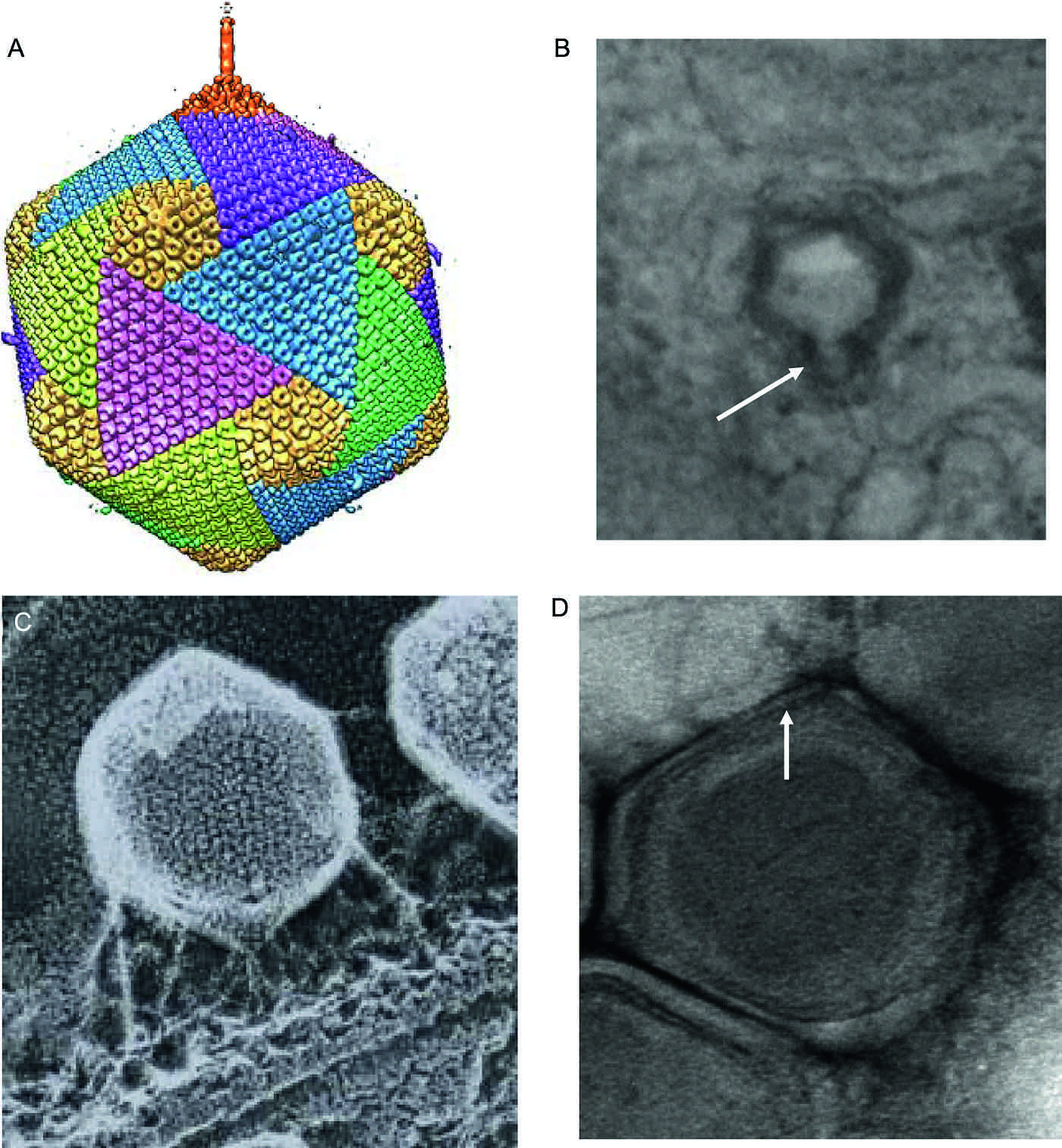
Figure 2 (Top) Genome organization of PBCV1. (Bottom) Circular map of the EsV-1 genome. Inner circle, sites for restriction endonucleases AscI and SfiI. Outer circle, nucleotide, coordinates and position of repeat regions (block rectangles: B, C, C, etc). Triangles, the inverted terminal repeats, ITRs A and A.
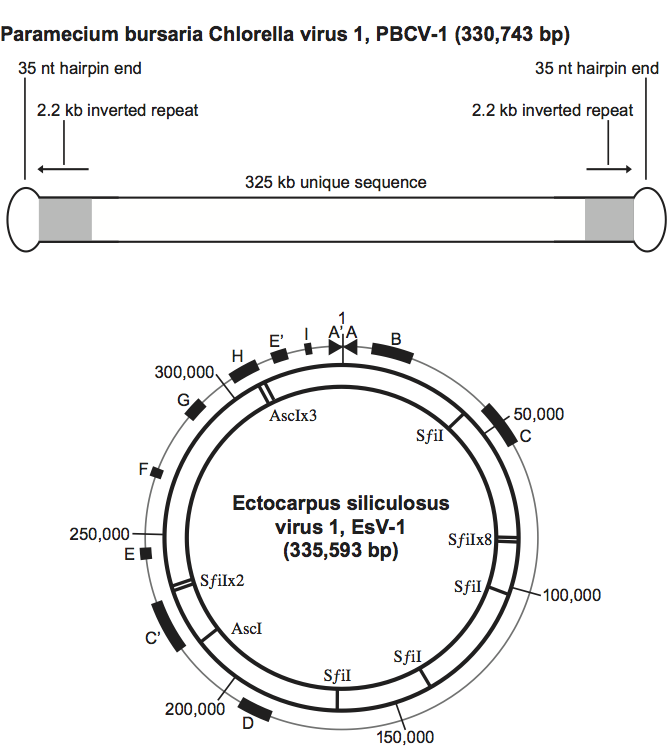
Figure 3 Schematic of the proposed life cycle of EhV-86. Enveloped EhV-86 enters E. huxleyi with an intact capsid and nucleoprotein core either by an endocytotic mechanism (step 1a) followed by fusion of its envelope with the vacuole membrane (step 2) or by fusion of its envelope with the host plasma membrane (step 1b). The viral capsid encapsulated nucleoprotein core rapidly targets the nucleus where capsid breakdown releases the viral genome (step 3). The viral genome enters the host nucleus where early promoter sequences are expressed by host RNA polymerase. Mid-late genes are expressed by viral RNA polymerase within the cytoplasm where capsid assembly takes place, possibly by filling of a pro-capsid with viral DNA and core proteins (step 4). Early assembled viruses are transported to the plasma membrane (step 5) where they are released by a budding mechanism (step 6)
(from Mackinder et al. (2010). J. Gen. Virol., 90(9), 2306-2316; with permission).
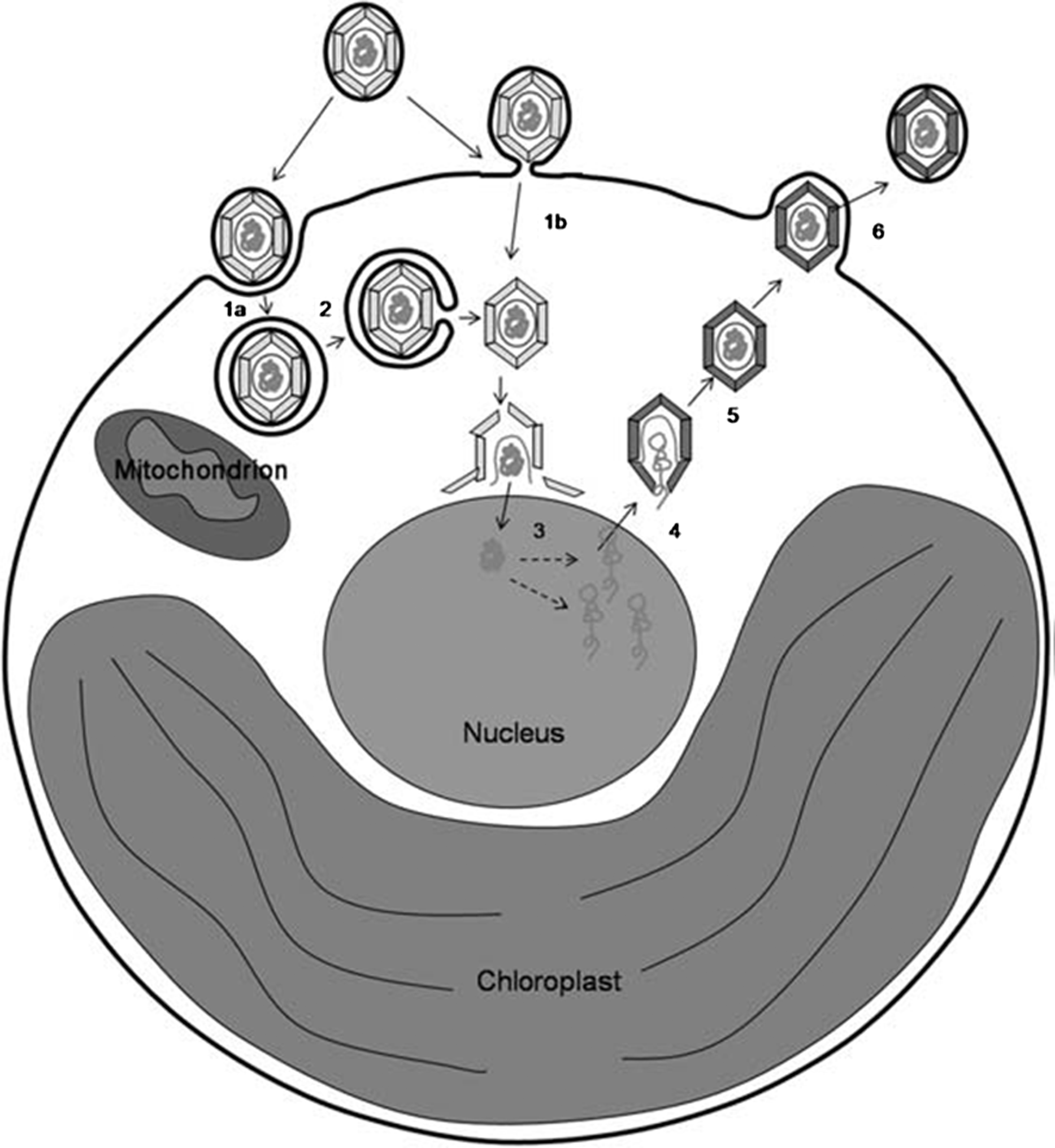
Figure 4 Phylogenetic analysis of members of the family Phycodnaviridae based on a distance matrix algorithm between the amino acid sequences of DNA pol fragments of phycodnaviruses and other large dsDNA viruses (Neighbor in PHYLIP, version 3.61). The alignment was performed (ClustalW) on the region spanning the highly conserved regions I and IV of the DNA pol genes. Abbreviations: Micromonas pusilla virus (MpV); Ostreococcus tauri virus (OtV); paramecium bursaria chlorella virus (PBCV); Emiliania huxleyi virus (EhV); Ectocarpus siliculosus virus (EsV); Feldmannia irregularis virus (FirrV); Feldmannia species virus (FsV), Chrysochromulina ericina virus (CeV); Pyramimonas orientalis virus (PoV); Chysochromulina brevifilum virus (CbV); Phaeocystis globosa virus (PgV); Heterosigma akashiwo virus (HaV); equid herpesvirus (EHV); alcelaphine herpesvirus (AlHV); bovine herpesvirus (BOHV); African swine fever virus (ASFV); Autographa californica multiple nucleopolyhedrovirus (AcMNPV); Bombyx mori nucleopolyhedrovirus (BmNPV); Lymantria dispar multiple nucleopolyhedrovirus (dMNDV); Helicoverpa zea single nucleopolyhedrovirus (HzSNPV); Chironomus luridus entomopoxvirus; (CLEV); vaccinia virus (VACV); fowlpox virus (FWPV); molluscum contagiosum virus (MOCV); invertebrate iridescent virus (IIV; family Iridoviridae). The scale bar indicates a distance of 0.2 fixed mutations per amino acid.
(Courtesy of Ilana Gilg.)