Family: Papillomaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are non-enveloped, 55 nm in diameter. The icosahedral capsid is composed of 72 capsomers in skewed (T=7) arrangement (Figure 1). Filamentous and tubular forms are observed as a result of aberrant maturation.
Physicochemical and physical properties
The virion Mr is 47 ×106. Buoyant density of virions in sucrose and CsCl gradients is 1.20 and 1.34–1.35 g cm−3, respectively. Virion S20,W is 300. Virions are resistant to ether, acid and heat treatment (50°C, 1 h).
Nucleic acid
Virions contain a single molecule of circular dsDNA. The genomic size ranges between 6800 and 8400 bp. The DNA constitutes about 10–13% of the virion by weight. The G+C content is 40–60%. In the mature virion, the viral DNA is associated with host cell histone proteins H2a, H2b, H3 and H4 in a chromatin-like complex.
Diagram of the bovine papillomavirus 1 (BPV-1) genome. The viral dsDNA (size in bp, origin of replication: ori) is indicated. The outer arrows indicate the protein-coding ORFs and their direction of transcription.
Proteins
The virus genomes encode 8–10 proteins with sizes ranging from 7 to 73 kDa (Table 1). L1 and L2 make up the capsid. E1 and E2 are involved in replication and in intragenomic regulation (E2). E5, E6 and E7 induce cellular DNA replication. E4 may represent a late function and binds to specific cytoskeleton structures. Genetic evidence has not been presented that associates specific viral proteins with the E3 and E8 ORFs.
Table 1 Deduced molecular masses of papillomavirus proteins (kDa)
| Virus | CRPV | BPV-1 | HPV-1 |
| Structural proteins |
|
|
|
| L1 | 57.9 | 55.5 | 59.6 |
| L2 | 52.8 | 50.1 | 50.7 |
| Nonstructural proteins |
|
|
|
| E1 | 67.9 | 68.0 | 73.0 |
| E2 | 44.0 | 48.0 | 41.8 |
| E4 | 25.8 | 12.0 | 10.4 |
| E5 | 11.3 | 7.0 | 9.4 |
| E6 | 29.7 | 15.1 | 19.2 |
| E7 | 10.5 | 14.0 | 11.0 |
Abbreviations: CRPV, cottontail rabbit papillomavirus; BPV, bovine papillomavirus; HPV, human papillomavirus.
Lipids
None present.
Carbohydrates
None present.
Genome organization and replication
Virions that attach to cellular receptors are engulfed by the cell, and the DNA is uncoated and transported to the nucleus. During productive infection, transcription of the viral genome is divided into early and late stages.
Transcription of the early and late ORFs occurs from the same strand in one direction only. Precursor mRNAs undergo post-transcriptional processing, which includes capping and polyadenylation of the 5’ and 3’ termini, respectively, as well as splicing. Efficient use of coding information involves differential splicing of the RNAs and utilization of overlapping ORFs. Early mRNAs encode regulatory proteins that may exhibit trans-activating properties. These include proteins that are required for DNA replication. Their expression leads to depression of some host cell enzymes and may also stimulate host cell DNA synthesis. Prior to the start of late events, viral DNA replication is initiated in the nucleus. Translation of the late transcripts produces structural proteins that are involved in capsid assembly. Post-translational modifications of some early and late viral proteins include phosphorylation, N-acetylation, ADP ribosylation and other events. Several of the viral proteins contain sequences, termed nuclear localization signals, which facilitate transport of the proteins to the host cell nucleus where virion maturation occurs. Virions are released by lysis of the virus-producing cells.
The genomes of most members of the family Papillomaviridae that have been sequenced contain 9–10 ORFs, labelled E1–E8 and L1–L2 (Figure 3). Some members lack the E3 and E8 ORFs. Proteins encoded by the E ORFs, with the possible exception of E4, represent non-structural polypeptides involved in transcription, DNA replication and transformation, whereas those encoded by the L ORFs represent structural proteins. Replication of the viral genome is initiated bi-directionally by specific binding of the E1 and E2 proteins at a unique origin of replication.
Antigenic properties
The L1 protein contains type-specific domains, and the L2 protein contains group-specific epitopes (see section on species demarcation for descriptions of groups and types). The availability of papillomavirus-like particles, resulting from the expression of L1 or L1 and L2 in baculovirus, vaccinia virus or yeast systems, has permitted a detailed analysis of antigenic characteristics.
Biological properties
Papillomaviruses are highly host species-specific and tissue-restricted. All known human papillomaviruses (HPVs) require terminal differentiation for replication and virion production. Infection appears to occur mainly via microlesions of proliferating basal layer cells. Except for inefficient replication of HPVs in raft cultures of human keratinocytes, or more efficiently in human skin or mucosal xenografts in immunocompromised rodents, HPV replication has not been achieved in cell culture systems.
Virus spread occurs by release of virions from the surface of warts and papillomatous lesions, which frequently contain large quantities of viral particles within the superficial differentiated layers. Virus reactivation is particularly frequent under conditions of immunosuppression. The mode of viral DNA persistence and possible clearance of HPV infections by immunological interference are still poorly investigated.
Transmission of viral infections occurs by close contacts. Papillomavirus types are distributed worldwide. They cause benign tumors (warts and papillomas) in their natural host and occasionally in related species. Frequently the infection leads to microlesions, which are barely or not at all visible without optical aid. Papillomas are induced in the skin and in mucous membranes, often at specific sites of the body. Some papillomatous proliferations induced by specific types of papillomaviruses bear a high risk for malignant progression. Specific human cancers (e.g. cervical carcinoma, anal, vulval and penile cancers, and specific squamous cell carcinomas of the skin) have been linked to certain types of HPV infection (e.g. by HPV-16 and HPV-18, HPV-5 and HPV-8, and several others). Viral DNA is often, but not always, present in an integrated form, particularly in cervical cancers, whereas skin carcinomas appear to harbor the viral genome in an episomal state. Cancer-linked anogenital HPV types efficiently immortalize a wide variety of human cells in tissue culture. Immortalization results from functions of the E6 and E7 genes, which act cooperatively, although both genes are able to immortalize human cells independently at low efficiency. E6 binds and degrades the cellular p53 protein and stimulates the telomerase enzyme, whereas E7 interacts with the cellular pRB and some related proteins, and directly activates cyclins E and A. Interactions of the viral oncoproteins with cellular cyclin-dependent kinase inhibitors (p16, p21 and p27) also emerge as important events in immortalization.
Species demarcation criteria in the family
The demarcation of papillomavirus species by phenotypic criteria similar to those applied to other families of viruses is problematic for a variety of reasons. One is that papillomaviruses do not elicit consistent humoral immune responses in infected human or other mammalian individuals, and it is therefore not possible to develop a taxonomy based on serology. The lack of reliable cell culture and laboratory animal host systems represents further limitations. Moreover, the coverage of different papillomaviruses in the scientific literature is very heterogenous, ranging from scattered single publications addressing individual, rare papillomaviruses to the body of 20,000 biological, medical and epidemiological publications addressing HPV-16 and HPV-18, the causes of cervical cancer. From the beginning of papillomavirus nomenclature in the 1930s, researchers were confronted with the problem of providing succinct names and distinguishing criteria for viruses that share many characteristics, such as similar genome sizes, similar target tissue properties (e.g. mucosal and cutaneous), and similar etiologies ranging from latent infections to various forms of neoplasia. In spite of these limitations, two principal pillars for papillomavirus taxonomy emerged. (1) All known papillomaviruses are strictly host species-specific, and this restriction needs to be reflected in the taxonomy. (2) DNA sequence comparisons led to refined phylogenetic studies, which show that all papillomavirus genomes are monophyletic in origin, that they evolve more slowly than virtually any other group of viruses, and that they do not recombine. The topology of phylogenetic trees is an indispensable criterion for taxonomic evaluation of this virus family.
From their roots nearly 80 years ago as recognized agents of disease, papillomaviruses have been described as “types”. This universal usage could potentially have led to the types being defined as species. However, the very large number of types prompted species to be set at a higher level, with the result that many species contain more than one type, with the species name derived from a prominent type in the species. The various species are also well supported by distinct biological properties. Similarly, genera were defined by phylogenetic considerations, relationships between host species and major differences in genome organization. More details on these issues, including the quantitative criteria utilized to define types, species and genera, are discussed in de Villiers et al. (2004) and Bernard et al. (2010). The latter paper contains extensive proposals to recognize additional species and genera, and to rationalize papillomavirus taxonomy generally. These proposals are currently under consideration by the ICTV.
Genus Alphapapillomavirus
Type species Human papillomavirus 32
Distinguishing features
Members of this genus preferentially infect the oral or anogenital mucosa in humans and primates. Members of certain species (e.g. Human papillomavirus 2 and Human papillomavirus 10) are also found in lesions of cutaneous sites. Members of some species (e.g. Human papillomavirus 16 and Human papillomavirus 18) are considered as high-risk in view of their regular presence in malignant tissue and their in vitro transforming activities. Members of other species (e.g. Human papillomavirus 53, Human papillomavirus 26 and Human papillomavirus 34) cause malignant or benign lesions, whereas low-risk viruses (in Human papillomavirus 61, Human papillomavirus 7, Human papillomavirus 6, Human papillomavirus 54, Human papillomavirus cand90 and Human papillomavirus 71) mainly cause benign lesions. Genome organization: an E5 ORF is conserved between the early and late coding regions.
List of species in the genus Alphapapillomavirus
| Human papillomavirus 2 |
|
|
| Human papillomavirus – 2 | [X55964] | (HPV-2) |
| Human papillomavirus – 27 | [X73373] | (HPV-27) |
| Human papillomavirus – 57 | [X55965] | (HPV-57) |
| Human papillomavirus – 6 |
|
|
| Human papillomavirus – 6 | [X00203] | (HPV-6) |
| Human papillomavirus – 11 | [M14119] | (HPV-11) |
| Human papillomavirus – 13 | [X62843] | (HPV-13) |
| Human papillomavirus – 44 | [U31788; U31791] | (HPV-44) |
| Human papillomavirus – 74 | [U40822] | (HPV-74) |
| Pygmy champanzee papillomavirus -1 | [X62844] | (PCPV-1) |
| Pygmy champanzee papillomavirus -1 - Champanzee | [AF020905] | (PCPV-1C) |
| Human papillomavirus 7 |
|
|
| Human papillomavirus – 7 | [X74463] | (HPV-7) |
| Human papillomavirus – 40 | [X74478] | (HPV-40) |
| Human papillomavirus – 43 | [AJ620205] | (HPV-43) |
| Human papillomavirus – cand91 | [AF131950] | (HPV-cand91) |
| Human papillomavirus 10 |
|
|
| Human papillomavirus – 3 | [X74462] | (HPV-3) |
| Human papillomavirus – 10 | [X74465] | (HPV-10) |
| Human papillomavirus – 28 | [U31783] | (HPV-28) |
| Human papillomavirus – 29 | [U31784] | (HPV-29) |
| Human papillomavirus – 77 | [Y15175] | (HPV-77) |
| Human papillomavirus 78 |
| (HPV-78) |
| Human papillomavirus – 94 | [AJ620021] | (HPV-94) |
| Human papillomavirus 16 |
|
|
| Human papillomavirus – 16 | [K02718] | (HPV-16) |
| Human papillomavirus – 31 | [J04353] | (HPV-31) |
| Human papillomavirus – 33 | [M12732] | (HPV-33) |
| Human papillomavirus – 35 | [X74476] | (HPV-35) |
| Human papillomavirus – 52 | [X74481] | (HPV-52) |
| Human papillomavirus – 58 | [D90400] | (HPV-58) |
| Human papillomavirus – 67 | [D21208] | (HPV-67) |
| Human papillomavirus 18 |
|
|
| Human papillomavirus – 18 | [X05015] | (HPV-18) |
| Human papillomavirus – 39 | [M62849] | (HPV-39) |
| Human papillomavirus – 45 | [X74479] | (HPV-45) |
| Human papillomavirus – 59 | [X77858] | (HPV-59) |
| Human papillomavirus – 68 | [X67161] | (HPV-68) |
| Human papillomavirus – 70 | [U21941] | (HPV-70) |
| Human papillomavirus – cand85 | [AF131950] | (HPV-cand85) |
| Human papillomavirus – 26 |
|
|
| Human papillomavirus – 26 | [X74472] | (HPV-26) |
| Human papillomavirus – 51 | [M62877] | (HPV-51) |
| Human papillomavirus – 69 | [AB027020] | (HPV-69) |
| Human papillomavirus – 82 | [AB027021] | (HPV-82) |
| Human papillomavirus - 32 |
|
|
| Human papillomavirus - 32 | [X74475] | (HPV-32) |
| Human papillomavirus - 42 | [M73236] | (HPV-42) |
| Human papillomavirus 34 |
|
|
| Human papillomavirus – 34 | [X74476] | (HPV-34) |
| Human papillomavirus – 73 | [X94165] | (HPV-73) |
| Human papillomavirus 53 |
|
|
| Human papillomavirus – 30 | [X74474] | (HPV-30) |
| Human papillomavirus – 53 | [X74482] | (HPV-53) |
| Human papillomavirus – 56 | [X74483] | (HPV-56) |
| Human papillomavirus – 66 | [U31794] | (HPV-66) |
| Human papillomavirus 54 |
|
|
| Human papillomavirus – 54 | [U37488] | (HPV-54) |
| Human papillomavirus – 61 | [U31793] | (HPV-61) |
| Human papillomavirus – 72 | [X94164] | (HPV-72) |
| Human papillomavirus – 81 | [AJ620209] | (HPV-81) |
| Human papillomavirus – 83 | [AF151983] | (HPV-83) |
| Human papillomavirus – 84 | [AF293960] | (HPV-84) |
| Human papillomavirus – cand62 | [U12499] | (HPV-cand62) |
| Human papillomavirus – cand86 | [AF349909] | (HPV-cand86) |
| Human papillomavirus – cand87 | [AJ400628] | (HPV-cand87) |
| Human papillomavirus – cand89 | [AF436128] | (HPV-cand89) |
| Human papillomavirus 71 |
|
|
| Human papillomavirus – 71 | [AB040456] | (HPV-71) |
| Human papillomavirus cand90 |
|
|
| Human papillomavirus – cand90 | [AY057438] | (HPV-cand90) |
| Rhesus monkey papillomavirus 1 |
|
|
| Rhesus monkey papillomavirus - 1 | [M60184] | (RhPV-1) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Alphapapillomavirus but have not been approved as species
None reported.
Genus Betapapillomavirus
Type species Human papillomavirus 5
Distinguishing features
Members of this genus preferentially infect the skin of humans. These infections are latent in the general population, but are activated under conditions of immunosuppression. Members of the species Human papillomavirus 5, Human papillomavirus 9 and Human papillomavirus 49 are also associated with the disease epidermodysplasia verruciformis. Genome organization: E5 ORF is absent.
List of species in the genus Betapapillomavirus
| Human papillomavirus 5 |
|
|
| Human papillomavirus – 5 | [M17463] | (HPV-5) |
| Human papillomavirus – 8 | [M12737] | (HPV-8) |
| Human papillomavirus – 12 | [X74466] | (HPV-12) |
| Human papillomavirus – 14 | [X74467] | (HPV-14) |
| Human papillomavirus – 19 | [X74470] | (HPV-19) |
| Human papillomavirus – 20 | [U31778] | (HPV-20) |
| Human papillomavirus – 21 | [U31779] | (HPV-21) |
| Human papillomavirus – 24 | [U31782] | (HPV-24) |
| Human papillomavirus – 25 | [U74471] | (HPV-25) |
| Human papillomavirus – 36 | [U31785] | (HPV-36) |
| Human papillomavirus – 47 | [M32305] | (HPV-47) |
| Human papillomavirus 9 |
|
|
| Human papillomavirus – 9 | [X74464] | (HPV-9) |
| Human papillomavirus – 15 | [X74468] | (HPV-15) |
| Human papillomavirus – 17 | [X74469] | (HPV-17) |
| Human papillomavirus – 22 | [U31780] | (HPV-22) |
| Human papillomavirus – 23 | [U31781] | (HPV-23) |
| Human papillomavirus – 37 | [U31786] | (HPV-37) |
| Human papillomavirus – 38 | [U31787] | (HPV-38) |
| Human papillomavirus – 80 | [Y15176] | (HPV-80) |
| Human papillomavirus 49 |
|
|
| Human papillomavirus – 49 | [X74480] | (HPV-49) |
| Human papillomavirus – 75 | [Y15173] | (HPV-75) |
| Human papillomavirus – 76 | [Y15174] | (HPV-76) |
| Human papillomavirus cand92 |
|
|
| Human papillomavirus – cand92 | [AF531420] | (HPV-cand92) |
| Human papillomavirus – cand96 |
|
|
| Human papillomavirus – cand96 | [AY382779] | (HPV-cand96) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Betapapillomavirus but have not been approved as species
None reported.
Genus Gammapapillomavirus
Type species Human papillomavirus 4
Distinguishing features
Members of this genus cause cutaneous lesions in their host and are histologically distinguishable by intracytoplasmic inclusion bodies that are species specific. Genome organization: E5 ORF is absent.
List of species in the genus Gammapapillomavirus
| Human papillomavirus 4 |
|
|
| Human papillomavirus – 4 | [X70827] | (HPV-4) |
| Human papillomavirus – 65 | [X70829] | (HPV-65) |
| Human papillomavirus – 95 | [AJ620210] | (HPV-95) |
| Human papillomavirus 48 |
|
|
| Human papillomavirus – 48 | [U31790] | (HPV-48) |
| Human papillomavirus 50 |
|
|
| Human papillomavirus – 50 | [U31790] | (HPV-50) |
| Human papillomavirus 60 |
|
|
| Human papillomavirus – 60 | [U31792] | (HPV-60) |
| Human papillomavirus 88 |
|
|
| Human papillomavirus – 88 |
| (HPV-88) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Gammapapillomavirus but have not been approved as species
None reported.
Genus Deltapapillomavirus
Type species European elk papillomavirus
Distinguishing features
These viruses induce fibropapillomas in their respective ungulate hosts. Trans-species transmission occurs, where it induces sarcoids. Genome organization: ORFs located in the region between the early and late genes have transforming properties.
List of species in the genus Deltapapillomavirus
| Bovine papillomavirus 1 |
|
|
| Bovine papillomavirus - 1 | [X02346] | (BPV-1) |
| Bovine papillomavirus - 2 | [M20219] | (BPV-2) |
| Deer papillomavirus | [M11910] | (DPV) |
| (Deer fibroma virus) |
|
|
| European elk papillomavirus |
|
|
| European elk papillomavirus | [M15953] | (EEPV) |
| Reindeer papillomavirus | [AF443292] | (RPV) |
| Ovine papillomavirus 1 |
|
|
| Ovine papillomavirus – 1 | [U83594] | (OvPV-1) |
| Ovine papillomavirus – 2 | [U83595] | (OvPV-2) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Deltapapillomavirus but have not been approved as species
None reported.
Genus Epsilonpapillomavirus
Type species Bovine papillomavirus 5
Distinguishing features
Infections cause cutaneous papillomas in cattle.
List of species in the genus Epsilonpapillomavirus
| Bovine papillomavirus 5 | ||
| Bovine papillomavirus 5 | [AF457465] | (BPV-5) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Epsilonpapillomavirus but have not been approved as species
None reported.
Genus Zetapapillomavirus
Type species Equine papillomavirus 1
Distinguishing features
Infections cause cutaneous lesions in horses. Genome organization: an ORF of unknown functional relevance overlaps with the L2 ORF.
List of species in the genus Zetapapillomavirus
| Equine papillomavirus 1 | ||
| Equus caballus papillomavirus 1 | [AF498323] | (EcPV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Zetapapillomavirus but have not been approved as species
None reported.
Genus Etapapillomavirus
Type species Fringilla coelebs papillomavirus
Distinguishing features
Avian papillomaviruses causing cutaneous lesions in their host. Genome organization: an ancestral E7 ORF exists which has partial E6 characteristics. Typical E6 ORF is absent.
List of species in the genus Etapapillomavirus
| Fringilla coelebs papillomavirus | ||
| Chaffinch papillomavirus | [AY957109] | (FcPV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Etapapillomavirus but have not been approved as species
None reported.
Genus Thetapapillomavirus
Type species Psittacus erithacus timneh papillomavirus
Distinguishing features
Avian papillomaviruses causing cutaneous lesions in their host. Genome organization: an ancestral E7 ORF exists which have partial E6 characteristics. Typical E4, E5 and E6 ORFs are absent.
List of species in the genus Thetapapillomavirus
| Psittacus erithacus timneh papillomavirus | ||
| Psittacus erithacus timneh papillomavirus | [AF420235] | (PePV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Thetapapillomavirus but have not been approved as species
None reported.
Genus Iotapapillomavirus
Type species Mastomys natalensis papillomavirus
Distinguishing features
Rodent papillomavirus causing cutaneous lesions. Genome organization: the E2 ORF is considerably larger than in other genera and the E5 ORF is absent.
List of species in the genus Iotapapillomavirus
| Mastomys natalensis papillomavirus | ||
| Mastomys natalensis papillomavirus | [U01834] | (MnPV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Iotapapillomavirus but have not been approved as species
None reported.
Genus Kappapapillomavirus
Type species Cottontail rabbit papillomavirus
Distinguishing features
Members of this genus cause cutaneous and mucosal lesions in rabbits. Genome organization: the E6 ORF is larger than in other genera. An uncharacterized E8 ORF is present in the early region.
List of species in the genus Kappapapillomavirus
| Cottontail rabbit papillomavirus | ||
| Cottontail rabbit papillomavirus | [K02708] | (CRPV) |
| Rabbit oral papillomavirus | ||
| Rabbit oral papillomavirus | [AF227240] | (ROPV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Kappapapillomavirus but have not been approved as species
None reported.
Genus Lambdapapillomavirus
Type species Canine oral papillomavirus
Distinguishing features
Members of this genus infect cats and dogs, causing mucosal and cutaneous lesions. Genome organization: the region between the early and late coding regions is exceptionally large, ranging between 1200 and 1500 bp.
List of species in the genus Lambdapapillomavirus
| Canine oral papillomavirus | ||
| Canine oral papillomavirus | [L22695] | (COPV) |
| Feline papillomavirus | ||
| Feline papillomavirus | [AF377865] | (FdPV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Lambdapapillomavirus but have not been approved as species
None reported.
Genus Mupapillomavirus
Type species Human papillomavirus 1
Distinguishing features
Human papillomaviruses causing cutaneous lesions in their host that are histologically distinguishable by species-specific intracytoplasmic inclusion bodies. Genome organization: the control region is larger than in other genera.
List of species in the genus Mupapillomavirus
| Human papillomavirus 1 | ||
| Human papillomavirus 1 | [V01116] | (HPV-1) |
| Human papillomavirus 63 | ||
| Human papillomavirus 63 | [X70828] | (HPV-63) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Mupapillomavirus but have not been approved as species
None reported.
Genus Nupapillomavirus
Type species Human papillomavirus 41
Distinguishing features
Human papillomaviruses causing benign and malignant cutaneous lesions. Genome organization: several larger ORFs are located in the L1 ORF region. The E2 binding sites in the control region all deviate from the typical consensus sequences, ACCGNNNNCGGT.
List of species in the genus Nupapillomavirus
| Human papillomavirus 41 | ||
| Human papillomavirus 41 | [X56147] | (HPV-41) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Nupapillomavirus but have not been approved as species
None reported.
Genus Xipapillomavirus
Type species Bovine papillomavirus 3
Distinguishing features
Infections cause true papillomas on the cutaneous or mucosal surfaces of cattle. Genome organization: a characteristic E6 ORF is absent and the E8 ORF located in this region displays transforming properties similar to that of viruses in the species Bovine papillomavirus 1.
List of species in the genus Xipapillomavirus
| Bovine papillomavirus 3 |
|
|
| Bovine papillomavirus 3 | [AF486184] | (BPV-3) |
| Bovine papillomavirus 4 | [X05817] | (BPV-4) |
| Bovine papillomavirus 6 | [AJ620208] | (BPV-6) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Xipapillomavirus but have not been approved as species
None reported.
Genus Omikronpapillomavirus
Type species Phocoena spinipinnis papillomavirus
Distinguishing features
These papillomaviruses have been isolated from genital warts in cetaceans. Genome organization: several larger ORFs are located in the L1 ORF region. True E7 ORF is absent.
List of species in the genus Omikronpapillomavirus
| Phocoena spinipinnis papillomavirus | ||
| Phocoena spinipinnis papillomavirus | [AJ238272] | (PsPV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Omikronpapillomavirus but have not been approved as species
None reported.
Genus Pipapillomavirus
Type species Hamster oral papillomavirus
Distinguishing features
Infections with these papillomaviruses cause mucosal lesions in hamsters. The E2 and L2 ORFs are partially overlapping.
List of species in the genus Pipapillomavirus
| Hamster oral papillomavirus | ||
| Hamster oral papillomavirus | [E15110] | (HaOPV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Pipapillomavirus but have not been approved as species
None reported.
List of other related viruses which may be members of the family Papillomaviridae but have not been approved as species
| Trichosurus vulpecula papillomavirus | [AF181682] | (TvPV) |
| Possum papillomavirus |
| (PoPV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
Putative papillomaviruses of a variety of different species have been identified from partial sequences. More than 300 such sequences are presently available in the databanks.
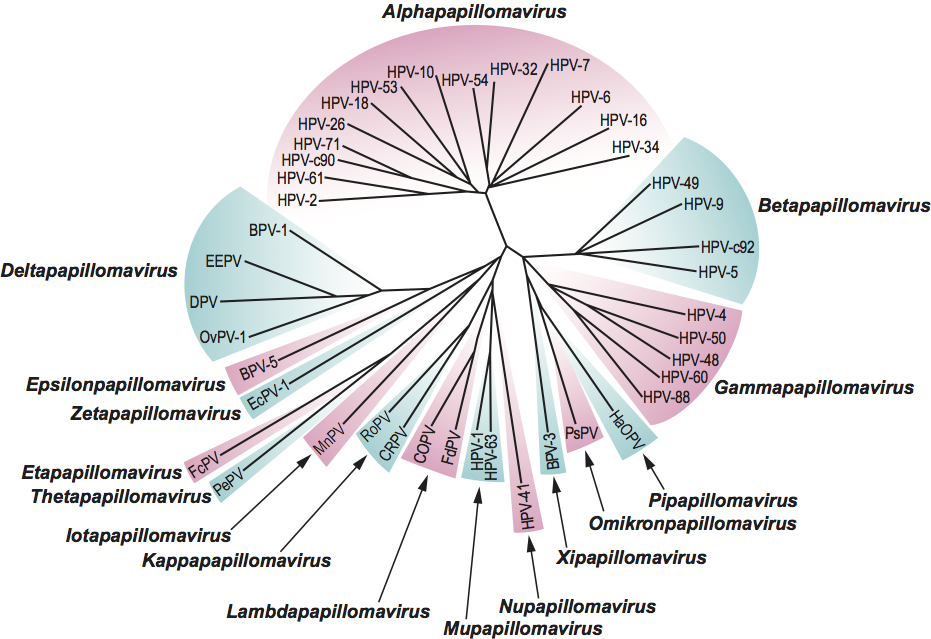
Phylogenetic relationships within the family
Phylogenetic relationships within the family are illustrated in Figure 4.
Similarity with other taxa
The families Papillomaviridae and Polyomaviridae share some similarities in morphology and nucleic acid composition, as well as in in vitro transforming activities of specific proteins.
Derivation of name
Papilloma: from Latin papilla, “nipple, pustule”, and Greek suffix -oma, used to form nouns denoting “tumors”.
Further reading
Bernard, H.U., Burk, R.D., Chen, Z., van Doorslaer, K., zur Hausen, H. and de Villiers, E.M. (2010). Classification of papillomaviruses based on 189 PV types and proposal of taxonomic amendments. Virology, 401, 70-79.
Bernard, H.U., Calleja-Macias, I.E. and Dunn, S.T. (2006). Genome variation of human papillomavirus types: Phylogenetic and medical implications. Intern. J. Cancer, 118, 1071-1076.
Campo, M.S. (Ed.) (2006). Papillomavirus Research. Caister Academic Press, Norfolk.
Chan, S.-Y., Delius, H., Halpern, A.L. and Bernard, H.-U. (1995). Analysis of genomic sequences of 95 papillomavirus types: Uniting typing, phylogeny and taxonomy. J. Virol., 69, 3074-3083.
de Villiers, E.-M. (1994). Human pathogenic papillomavirus types: an update. Curr. Top. Microbiol. Immunol., 186, 1-12.
de Villiers, E.M., Fauquet, C., Broker, T.R., Bernard, H.U. and zur Hausen, H. (2004). Classification of papillomaviruses. Virology, 324, 17-27.
Garcea, R.L. and DiMaio, D. (eds). (2007). The Papillomaviruses. Springer, Heidelberg.
Myers, G., Baker, C., Munger, K., Sverdrup, F., McBride, A., Bernard, H.-U. and Meissner, J. (Eds.) (1994–1997). Human Papillomaviruses, HPV Sequence Database, Volume 1–4. Los Alamos National Laboratory, Los Alamos, http://www.stdgen.lanl.gov/.
van Ranst, M., Kaplan, J.B. and Burk, R.D. (1992). Phylogenetic classification of human papillomaviruses: correlation with clinical manifestations. J. Gen Virol., 73, 2653-2660.
zur Hausen, H. (2002). Papillomavirus and cancer: from basic studies to clinical application. Nat. Rev. Cancer, 2, 342-350.
Contributed by
Bernard, H.-U., Burk, R.D., deVilliers, E.-M. and zur Hausen, H.
Figures
Figure 1 (Left) Atomic rendering of a papillomavirus capsid. Derived from an image reconstruction from electron cryomicroscopy of bovine papillomavirus (BPV) at 9 resolution combined with coordinates from the crystal structure of small virus-like particles of the human papillomavirus 16 (HPV-16) L1 protein (from Modis et al. (2002). EMBO J., 21, 47544762). (Centre) Schematic diagram representing the 72 capsomers in a T=7 arrangement of a papillomavirus capsid. The icosahedral structure includes 360 VP1 subunits arranged in 12 pentavalent and 60 hexavalent capsomers. (Right) Negative contrast electron micrograph of human papillomavirus 1 (HPV-1) virions. The bar represents 100 nm.
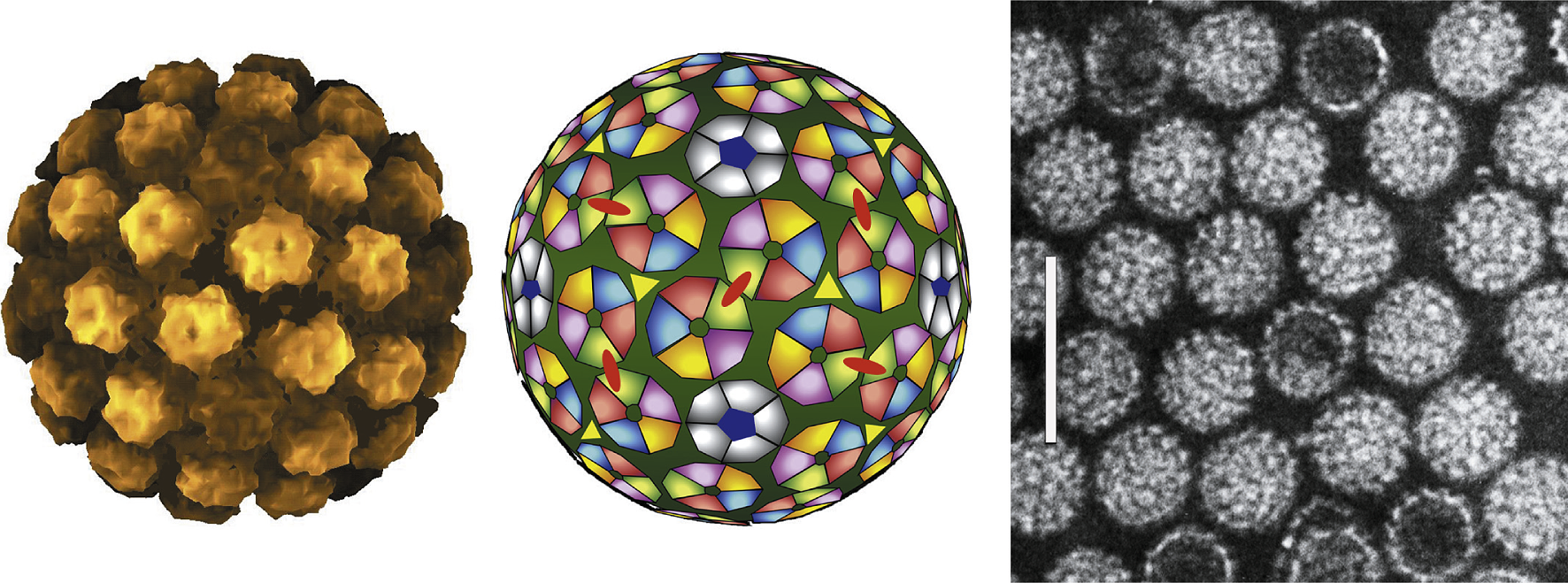
Figure 2 Diagram of the bovine papillomavirus 1 (BPV-1) genome. The viral dsDNA (size in bp, origin of replication: ori) is indicated. The outer arrows indicate the protein-coding ORFs and their direction of transcription.
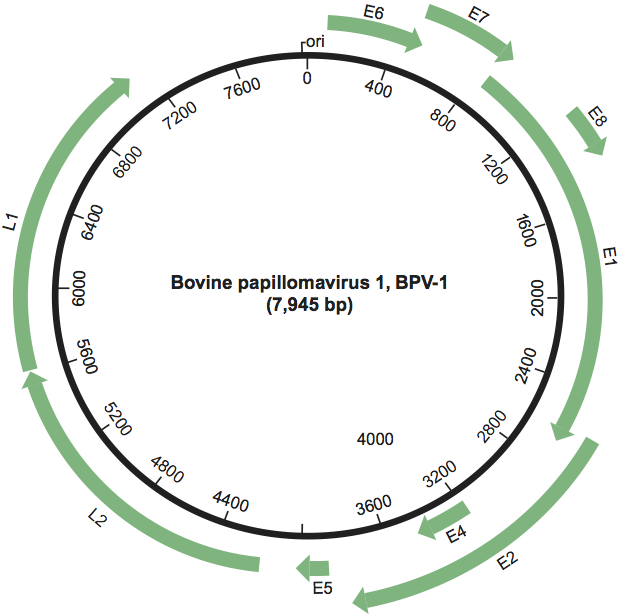
Figure 3 Comparison of genome organization for the viruses corresponding to the type species of each genus in the family Papillomaviridae. The circular dsDNA genomes have been linearized, with ori as the opening site (see Figure 2). Similar ORFs are indicated in similar colors. For abbreviations, see species lists in text.
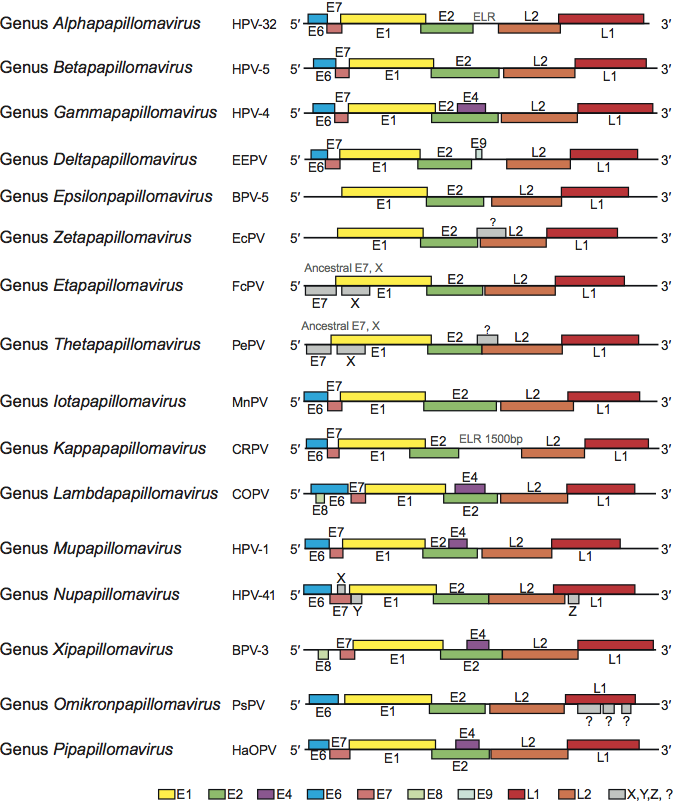
Figure 4 Phylogenetic tree representing the sequences of 118 papillomaviruses. The phylogenetically informative region of the L1 ORF was used in a modified version of the Phylip version 3.572 and based on a weighted version of the neighbor-joining analysis. Accession numbers are listed in the tables. The tree was constructed using the Treeview program (R. Page, University of Glasgow).