Family: Nimaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Whispovirus
Type species White spot syndrome virus
Virion properties
Morphology
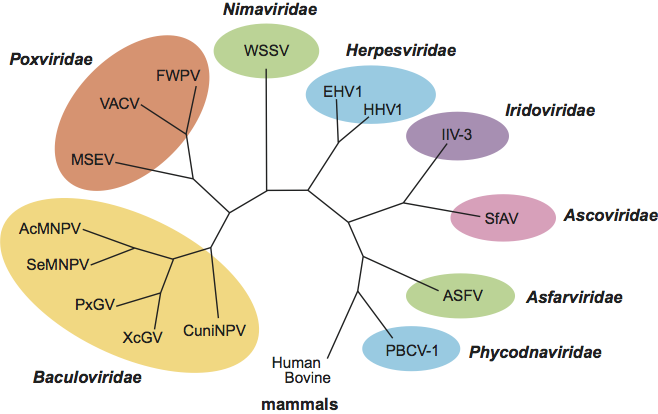
Virions are ovoid or ellipsoid to bacilliform in shape, have a regular symmetry, and measure 120–150 nm in diameter and 270–290 nm in length. Most notable is the thread- or flagellum-like extension (appendage) at one end of the virion. The virion consists of an inner, rod-shaped nucleocapsid with a tight-fitting capsid layer, an intermediate tegument layer and an outer lipid-containing trilaminar envelope. The isolated nucleocapsid typically measures 65–70 nm in diameter and 300–350 nm in length. It contains a DNA-protein core bounded by a distinctive capsid layer, giving it a cross-hatched or striated appearance (Figure 1).
Physicochemical and physical properties
Virions have a buoyant density of 1.22 g cm−3 in CsCl, whereas the nucleocapsids have a buoyant density of 1.31 g cm−3. The virions are sensitive to detergents. Other properties are not known.
Nucleic acid
The nucleocapsid contains a single molecule of circular dsDNA with an approximate size of 300 kbp. The G+C ratio of white spot syndrome virus (WSSV) is about 41%.
Proteins
The virions contain at least six major and at least 35 medium or minor polypeptides ranging in size from 14 to 664 kDa. The three structural elements of the virion each contain two major proteins: VP28 and VP19 in the envelope, VP26 and VP24 in the tegument, and VP664 and VP15 in the nucleocapsid. VP15 is a very basic, histone-like DNA-binding nucleoprotein. The three major proteins, VP28, VP26 and VP24, are phylogenetically related.
Lipids
Lipids are present in the envelope. The composition of lipids is known for virions purified from the crayfish Procambarus clarkii. The lipids are of host origin and are derived from host-cell nuclei.
Carbohydrates
None of the major virion structural proteins is glycosylated.
Genome organization and replication
The WSSV genome has, depending on the isolate, about 181 non-overlapping ORFs and almost equally distributed over both DNA strands (WSSV-CN; Figure 2). Only a few genes (20%) have been identified. These genes encode virion proteins (see below), proteins involved in DNA replication (DNA polymerase, ribonucleotide reductase subunits, dUTPase, thymidylate synthase, thymidine-thymidylate kinase) and protein-modifying proteins (protein kinase). The virion protein genes are not clustered, but are dispersed on both strands of the WSSV genome. The WSSV genome includes a very large gene (vp664) that encodes a major nucleocapsid protein of about 664 kDa. Most ORFs are unassigned. WSSV has also been shown to contain genes homologous to Marsupenaeusjaponicus and to Penaeus monodon. These may exemplify mimicry of the structure and function of host genes, e.g. to avoid detection and destruction by the host immune system, a stratagem used by other dsDNA viruses. Alternatively they may be degenerated proviral remnants.
The WSSV genome is further characterized by the presence of nine homologous repeat regions (Hr1-9; Figure 2). The number of imperfect palindromic repeats (250 bp in size) within an hr varies per isolate.
Viral transcripts are often but not always polyadenylated and they are usually capped. There is no evidence to suggest the occurrence of RNA splicing. Three immediate early genes have been identified in WSSV, and WSSV early genes and one of the immediately early genes has a promoter motif (-[a,c][a,c]TCAxT-) that matches the RNA polII core promoter of the Arthropods. The WSSV late genes’ consensus promoter (-A[a,t][a,t,g]AC-) is usually less than 100 nt (nucleotides) from the transcriptional start site. Consensus TATA box sequences have been found at about 25 bp upstream of early gene transcription initiation sites. Late transcripts seem to start 25 nt downstream of an A/T-rich region. Structural protein genes widely use polycistronic mRNAs and internal ribosome entry site elements to regulate their tranlsation.
Replication of WSSV occurs in the nucleus, where the virions are also assembled (Figure 1).
Genotypic variants exist that can be distinguished by restriction length polymorphism and on the basis of genomic sequence. Three WSSV isolates have been sequenced and they vary in size from 297,967 nt (WSSV-TH) to 305,107 nt (WSSV-CN) and 307,287 nt (WSSV-TW). Major differences can be attributed to deletions in WSSV-TH between 275,238 and 287,285, and between 267,203 and 268,046 and to the insertion of a 1.3 kb transposase sequence in WSSV-TW at 204,978–204,979 (Figure 2).
Antigenic properties
Polyclonal and monoclonal antibodies have been raised against WSSV virions, and they can be used as diagnostic tools. In recent years, researchers have found that the virus infection can be neutralized with antisera against the envelope proteins VP28, VP31, VP33 (VP36B) and VP12B (VP68). From this, it can be inferred that these proteins are involved in WSSV infection.
Biological properties
Host range
WSSV can infect a wide range of aquatic crustaceans including salt, brackish and fresh water penaeids, crabs and crayfish.
Transmission
The virus is transmitted by mouth by cannibalism of diseased individuals or via water through the gills. The virus can also be transmitted vertically from adults to offspring, either by being released from non-viable WSSV-infected eggs or from the supporting cells in ovarian tissue.
Geographical distribution
WSSV has been identified from crustaceans in China, Japan, Korea, Southeast Asia, South Asia, the Middle East and the Americas. An unusual property of the virus is the extremely wide host range despite a very low level of genetic polymorphism. The latter suggests that white spot disease (WSD) is a relatively recent epizootic.
Cytopathic effects
The major targets of WSSV infection are tissues of ectodermal and mesodermal embryonic origin such as subcuticular epithelial cells (including those of the shrimp stomach), gills, lymphoid organ and connective tissue. Although WSSV infects the underlying connective tissue in the shrimp hepatopancreas and midgut, the columnar epithelial cells of these two organs are of endodermal origin, and they do not become infected. WSSV can grow in primary cultures of lymphoid organ and ovary. Infection sometimes causes disease and sometimes not, depending upon factors as yet poorly understood but related to species tolerance and environmental triggers. Shrimp and other crustaceans susceptible to disease from this virus show gross signs of lethargy, such as lack of appetite and slow movement, and often a reddish coloration of the whole body together with “white spots” embedded within the exoskeleton. These spots are the result of calcified deposits that range in size from a few millimeters to 1 or more centimeters in diameter. However, in some cases such as acute experimental infections resulting from injected viral preparations, there may be no gross signs of infection other than lethargy and lack of appetite. Most affected animals die within 3–10 days after infection. With an appropriate infection dose to allow sufficient time before mortality, animals susceptible to disease show large numbers of virions circulating in the hemolymph, but this may also occur for tolerant species that show no mortality. Thus, high viral loads per se do not cause disease or mortality for all susceptible species.
Species demarcation criteria in the genus
Only a single species within the genus has been identified to date (White spot syndrome virus). Various isolates with small genetic polymorphisms have been identified (variants). It should be realized, however, that as the Nimaviridae is a newly recognized family, the species concept is subject to change after existing and new isolates have been studied in more detail.
List of species in the genus Whispovirus
| White spot syndrome virus |
|
|
| White spot syndrome virus - CN | [AF332093] | (WSSV-CN) |
| White spot syndrome virus -TH | [AF369029] | (WSSV-TH) |
| White spot syndrome virus -TW | [AF440570] | (WSSV-TW) |
| White spot syndrome virus |
|
|
| Chinese baculo-like virus |
|
|
| Hypodermic and hemotopoietic necrosis baculovirus |
|
|
| Penaeus monodon non-occluded baculovirus II |
|
|
| Penaeus monodon non-occluded baculovirus III |
|
|
| Penaeus monodon rod-shaped nuclear virus |
|
|
| Systemic ectodermal and mesodermal baculovirus |
|
|
| White spot bacilliform virus |
|
|
| White spot baculovirus |
|
|
Species names are in italic script; names of strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Whispovirus but have not been approved as species
None reported.
List of unassigned species in the family Nimaviridae
None reported.
Phylogenetic relationships within the family
Not applicable.
Similarity with other taxa
Morphologically, the WSSV virions and rod-shaped nucleocapsids resemble the budded virus particles of the baculoviruses, polydnaviruses and the unassigned Oryctes rhinoceros virus. The presence of repeat regions dispersed over the DNA genome is a property shared with members of the families Baculoviridae and Ascoviridae, the proposed Hytrosaviridae family and the unassigned nudiviruses. WSSV is phylogenetically distinct from other large DNA viruses (Figure 3).
Derivation of name
Nima: Latin for “thread”; referring to the thread- or tail-like polar extension (appendage) on the virus particle.
Further reading
Chou, H., Huang, C., Wang, C., Chiang, H., Lo, C., Chou, H.Y., Huang, C.Y., Wang, C.H., Chiang, H.C. and Lo, C.F. (1995). Pathogenicity of a baculovirus infection causing White spot syndrome in cultured penaeid shrimp in Taiwan. Diseas. Aquat. Org., 23, 165-173.
Durand, S., Lightner, D.V., Redman, R.M. and Bonami, J.R. (1997). Ultrastructure and morphogenesis of White spot syndrome baculovirus (WSSV). Diseas. Aquat. Org., 29, 205-211.
Huang, C., Zhang, X., Lin, Q., Xu, X., Hu, Z.H. and Hew, C.L. (2002). Proteomic analysis of shrimp White spot syndrome virus proteins and characterization of a novel envelope protein VP466. Molec. Cell. Proteom., 1, 223-231.
Leu, J.H., Yang, F., Zhang, X., Xu, X., Kou, G.H. and Lo, C.F. (2009). Whispovirus. Curr. Top. Microbiol. Immunol., 328, 197-227.
Lo, C.F., Ho, C.H., Chen, C.H., Liu, K.F., Chiu, Y.L., Yeh, P.Y., Peng, S.E., Hsu, H.C., Liu, H.C., Chang, C.F., Su, M.S., Wang, C.H. and Kou, G.H. (1997). Detection and tissue tropism of White spot syndrome baculovirus (WSBV) in captured brooders of Penaeus monodon with a special emphasis on reproductive organs. Diseas. Aquat. Org., 30, 53-72.
Nadala, E.C.B. and Loh, P.C. (1998). A comparative study of three different isolates of white spot virus. Diseas. Aquat. Org., 33, 231-234.
Tsai, J.M., Wang, H.C., Leu, J.H., Wang, A.H., Zhuang, Y., Walker, P.J., Kou, G.H. and Lo, C.F. (2006). Identification of the nucleocapsid, tegument, and envelope proteins of the shrimp white spot syndrome virus virion. J. Virol., 80, 3021-3029.
van Hulten, M.C.W., Westenberg, M., Goodall, S.D. and Vlak, J.M. (2000). Identification of two major virion protein genes of White spot syndrome virus of shrimp. Virology, 266, 227-236.
van Hulten, M.C.W., Witteveldt, J., Peters, S., Kloosterboer, N., Tarchini, R., Fiers, M., Sandbrink, H., Klein Lankhorst, R. and Vlak, J.M. (2001). The white spot syndrome virus DNA genome sequence. Virology, 286, 7-22.
Yang, F., He, J., Lin, X., Li, Q., Pan, D., Zhang, X. and Xu, X. (2001). Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol., 75, 11811-11820.
Contributed by
Lo, C.F., Aoki, T., Bonami, J.R., Flegel, T., Leu, J.H., Lightner, D.V., Stentiford, G., Söderhäll, K., Walker, P.J., Wang, H. C., Xun, X., Yang, F. and Vlak, J.M.
Figures
Figure 1 (Top) Morphology of virions of white spot syndrome virus (WSSV). (Left) Schematic illustration of the structure of a typical whispovirus virion. (Top center and right) Negative contrast electron micrographs of WSSV virions (center, courtesy of Marielle van Hulten) and nucleocapsids (right, courtesy of Don Lighter) from hemolymph of infected Penaeus monodon. The bars represent 100 nm. (Bottom left) Thin section of WSSV-infected stomach epithelium showing the parallel arrangement of virions in the nucleoplasm (the bar represents 1 m) and (bottom right) a cross-section of virions (the bar represents 1 m)
(courtesy of Don Lightner).
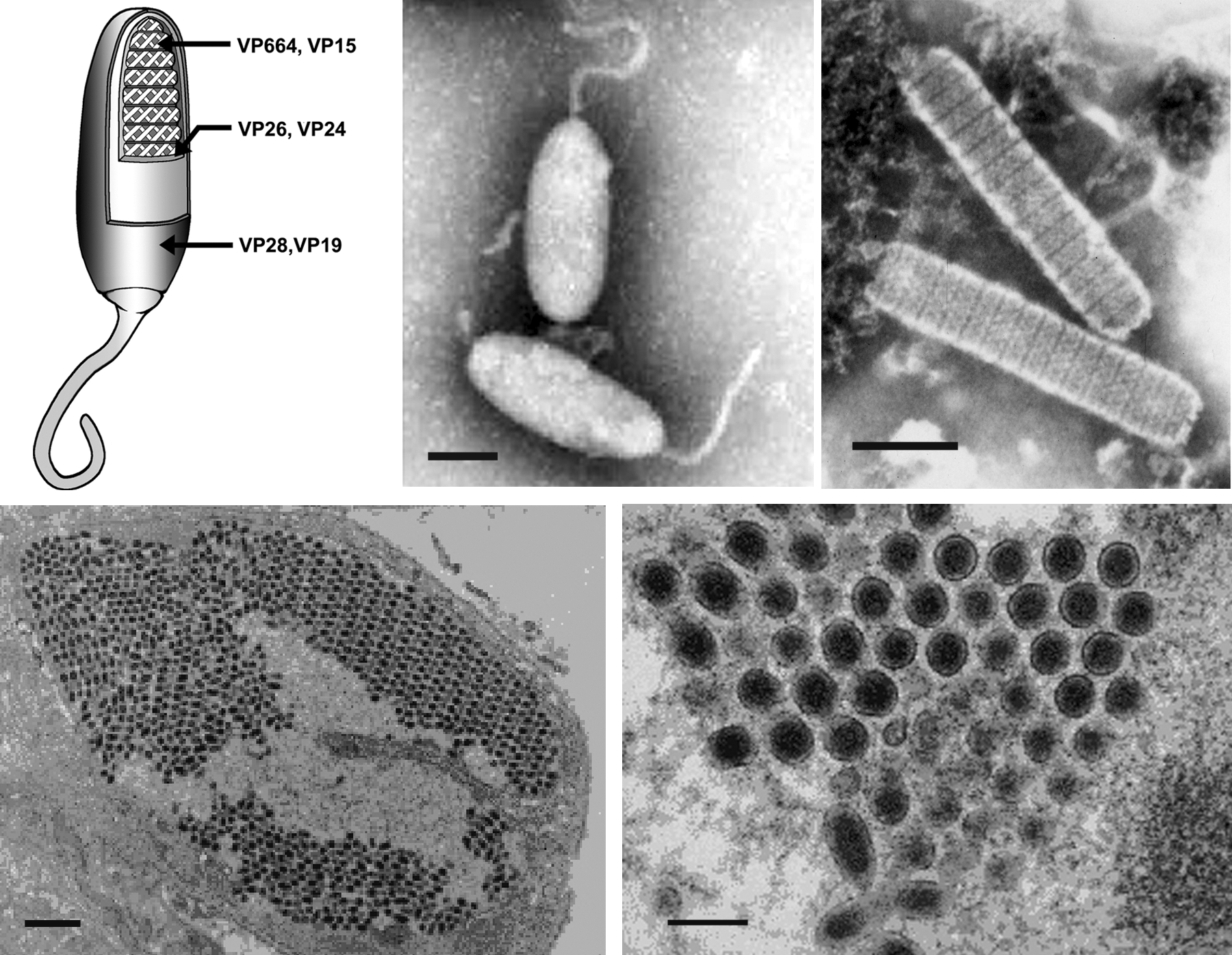
Figure 2 Schematic map of the circular dsDNA white spot syndrome virus type strain CN (WSSV-CN) genome showing the genomic organization. The solid arrows indicate the positions and direction of transcription of corresponding genes. The G at the start (GGATCC) of the largest BamHI fragment is designated position 1.
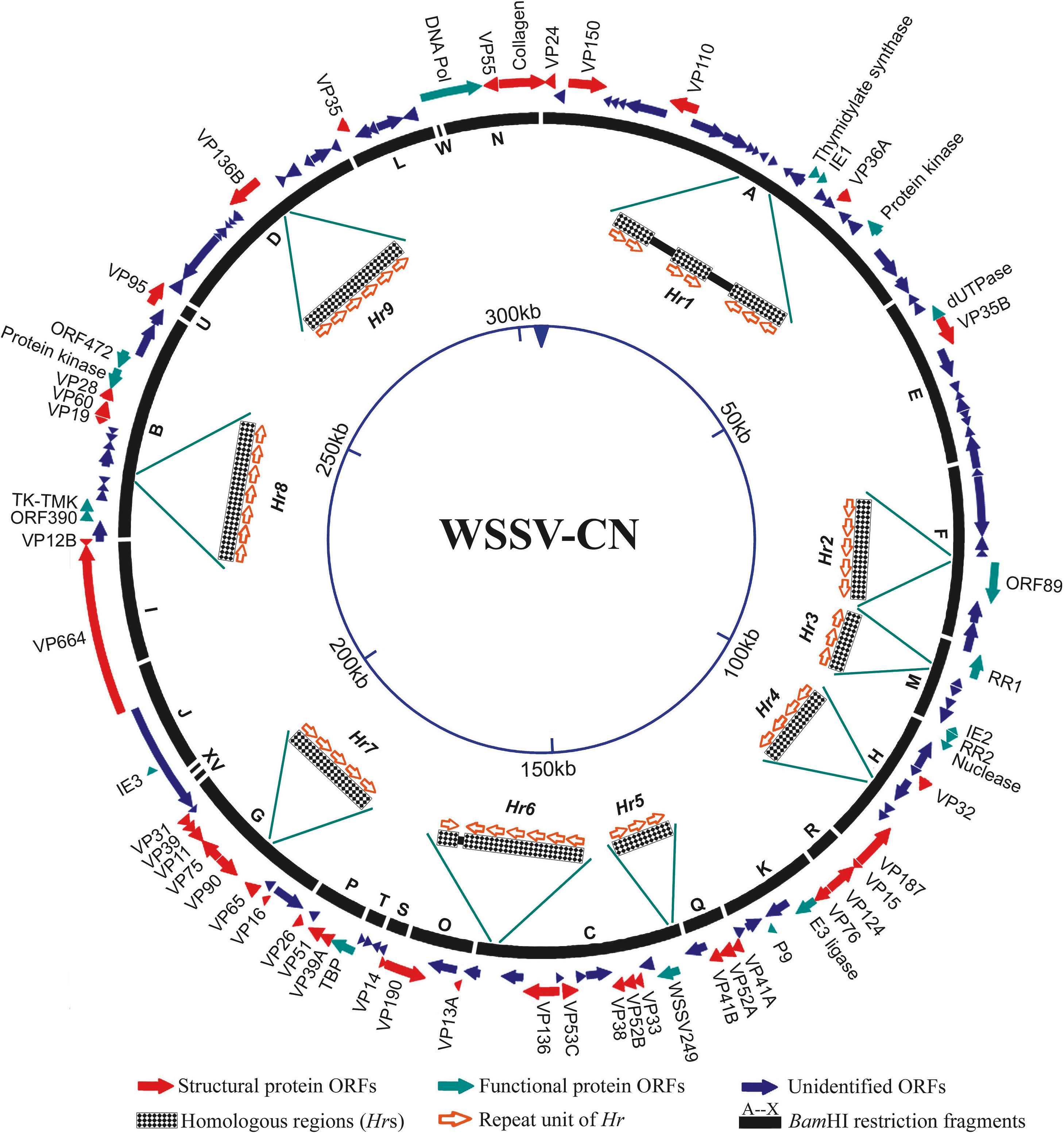
Figure 3 Cladogram based on genetic distances between aa sequences derived from the DNA polymerase gene of WSSV and a number of other large DNA viruses, as well as two mammalian representatives.