Family Myoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Distinguishing features of the family
Tails are contractile, more or less rigid, long and relatively thick (80–455×16–20 nm). They consist of a central core built of stacked rings of six subunits and surrounded by a helical contractile sheath, which is separated from the head by a neck. During contraction, sheath subunits slide over each other and the sheath becomes shorter and thicker. This brings the tail core in contact with the bacterial plasma membrane and is an essential stage of infection. Heads and tails are assembled in separate pathways. Compared to other tailed phages, myoviruses often have larger heads and higher particle weights and DNA contents, and seem to be more sensitive to freezing and thawing and to osmotic shock. Genera are differentiated by genome organization, mechanisms of DNA replication, and packaging, and the presence or absence of unusual bases and DNA polymerases.
Genus “T4-like viruses”
Type species Enterobacteria phage T4
Distinguishing features of the genus
Virions have elongated heads and tails with long, kinked fibers. Double stranded DNA genomes are circularly permuted and terminally redundant, and typically code for hydroxymethylcytosine synthesizing enzymes and B-type DNA polymerase. The genome is linear but circularly permuted and the DNA is packaged by a headful mechanism.
Virion properties
Morphology
Phage heads are prolate icosahedra (elongated pentagonal bipyramidal antiprisms), measure about 111×78 nm, and consist of 152 capsomers (T=13, elongated). Tails measure 113×16 nm and have a collar, base plate, six short spikes and six long fibers (Figure 1). Aberrant head structures (polyheads and isometric heads) are frequent.
Physicochemical and physical properties
Virion Mr is about 210 ×106, buoyant density in CsCl is 1.50 g cm−3, and S20,w about 1030S. Infectivity is ether and chloroform resistant.
Nucleic acid
Genomes have a Mr about 120 ×106, corresponding to 48% of the particle weight. DNA contains 5-hydroxymethylcytosine (HMC) instead of cytosine (these nucleotides are glycosylated), a G+C content of 35%, and is circularly permuted and terminally redundant. The Enterobacteria phage T4 (T4) genome has been sequenced (168,903 bp).
Proteins
T4 virions contain at least 49 proteins (8–155 kDa), including 1600–2000 copies of the major CP (43 kDa) and three proteins located inside the head. Various enzymes are encoded on the viral genome, e.g. type B (E. coli Pol II) DNA polymerase, numerous nucleotide metabolism enzymes and lysozyme (Figure 1).
Lipids
None reported.
Carbohydrates
Glucose is covalently linked to HMC in phage DNA.
Genome organization and replication
The genome is linear and comprises about 300 genes. Morphopoietic genes generally cluster together, but this is not true for all cases, suggesting extensive translocation of genes during evolution. The genome is circularly permuted and has 1–3% terminal redundancy. For this reason the genetic map is usually represented as a circle (Figure 2). After infection, the host chromosome breaks down and viral DNA replicates as a concatemer, generating forked replicative intermediates from multiple origins of replication. Transcription is regulated in part by phage-induced modification of host RNA polymerase and proceeds in three temporal waves (early, middle, late). Heads, tails and tail fibers are assembled in three separate pathways. Unique DNA molecules are packaged by a headful mechanism. Virions are assembled at the cell periphery.
Antigenic properties
A group antigen and antigens defining eight subgroups have been identified by complement fixation.
Biological properties
Phages are virulent, and infect enteric and related bacteria (Gammaproteobacteria). Their distribution is worldwide.
Species demarcation criteria in the genus
Species differ in host range, capsid size, serological properties and, insofar as known, DNA sequences.
List of species in the genus “T4-like viruses”
| Acinetobacter phage 133 |
|
|
| Acinetobacter phage 133 |
| (133) |
| Aeromonas phage 40RR2.8t |
|
|
| Aeromonas phage 40RR2.8t | [AY375531] | (40RR2.8t) |
| (Aeromonas phage 40R) |
| (40R) |
| Aeromonas phage 65 |
|
|
| Aeromonas phage 65 |
| (65) |
| Aeromonas phage Aeh1 |
|
|
| Aeromonas phage Aeh1 | [AY266303] | (Aeh1) |
| Enterobacteria phage SV14 |
|
|
| Enterobacteria phage D2A |
| (D2A) |
| Enterobacteria phage D8 |
| (D8) |
| Enterobacteria phage SV14 |
| (SV14) |
| Enterobacteria phage T4 |
|
|
| Enterobacteria phage C16 |
| (C16) |
| Enterobacteria phage F10 |
| (F10) |
| Enterobacteria phage Fs-alpha |
| (Fs-alpha) |
| Enterobacteria phage PST |
| (PST) |
| Enterobacteria phage SKII |
| (SKII) |
| Enterobacteria phage SKV |
| (SKV) |
| Enterobacteria phage SKX |
| (SKX) |
| Enterobacteria phage SV3 |
| (SV3) |
| Enterobacteria phage T2 |
| (T2) |
| Enterobacteria phage T4 | [AF158101] | (T4) |
| Enterobacteria phage T6 |
| (T6) |
| Pseudomonas phage 42 |
|
|
| Pseudomonas phage 42 |
| (42) |
| Vibrio phage nt-1 |
|
|
| Vibrio phage KVP20 |
| (KVP20) |
| Vibrio phage KVP40 | [AY283928] | (KVP40) |
| Vibrio phage nt-1 |
| (nt-1) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “T4-like viruses” but have not been approved as species
| Acinetobacter phage E4 |
| (E4) |
| Acinetobacter phage E5 |
| (E5) |
| Aeromonas phage 1 |
| (Aer1) |
| Aeromonas phage 25 |
| (25) |
| Aeromonas phage 31 |
| (31) |
| Enterobacteria phage 1 (Phage aeI) |
| (aeI) |
| Enterobacteria phage 11F |
| (11F) |
| Enterobacteria phage 3 |
| (3) |
| Enterobacteria phage 3T+ |
| (3T+) |
| Enterobacteria phage 50 |
| (50) |
| Enterobacteria phage 5845 |
| (5845) |
| Enterobacteria phage 66F |
| (66F) |
| Enterobacteria phage 8893 |
| (8893) |
| Enterobacteria phage 9/0 |
| (9/0) |
| Enterobacteria phage alpha1 |
| (alpha1) |
| Enterobacteria phage DdVI |
| (DdV1) |
| Enterobacteria phage F7 |
| (F7) |
| Enterobacteria phage Kl3 |
| (K13) |
| Enterobacteria phage RB42 |
| (RB42) |
| Enterobacteria phage RB43 |
| (RB43) |
| Enterobacteria phage RB49 |
| (RB49) |
| Enterobacteria phage RB69 | [AY303349] | (RB69) |
| Enterobacteria phage SMB |
| (SMB) |
| Enterobacteria phage SMP2 |
| (SMP2) |
Genus “P1-like viruses”
Type species Enterobacteria phage P1
Distinguishing features of the genus
Virions produce head size variants. DNA is circularly permuted and terminally redundant, and is packaged from a pac site. The genome is linear, and phages can carry out generalized transduction. Prophages persist as plasmids.
Virion properties
Morphology
Virions have icosahedral heads about 85 nm in diameter; head size variants (ca. 47–65 nm) have been observed. Tails measure 228×18 nm in Enterobacteria phage P1 (P1) and vary in length from 170 to 240 nm in other members of the genus (i.e., Enterobacteria phage P1D (P1D) and Aeromonas phage 43 (43)). Tails have base plates and six 90 nm-long kinked fibers. Particles with contracted tails aggregate side-by-side by means of exposed tail cores.
Physicochemical and Physical Properties
Phage P1 virion buoyant density is 1.48 g cm−3.
Nucleic Acid
Genomes comprise about 100 kbp and have a G+C content of 46%.
Proteins
Virions contain 24–28 constitutive proteins (10–220 kDa), including a major coat protein of 44 kDa.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genome is linear and carries about 100 genes; related functions are often distributed over several genome regions. Prophage DNA is circular. The genome is circularly permuted and terminally redundant (8–12%), and includes a recombinational hot spot (lox-cre). The genome also has an invertible tail fiber segment of about 4 kbp (C-loop) that is homologous to the G-loop of Enterobacteria phage Mu (Mu). Virion DNA circularizes after injection. Replication starts at a single site and has a phase of theta replication and then a phase of sigma structures, suggesting a rolling-circle mechanism. Progeny DNA is cut from concatemers at a pac site.
Antigenic properties
Phages P1, P2 and Mu share tail fiber antigens.
Biological properties
Phages are temperate, can carry out generalized transduction, and infect enteric and related gram-negative bacteria. Prophages are maintained as plasmids (1–2 copies per cell) or integrate (rarely) at specific sites into the bacterial chromosome. Prophages are weakly UV-inducible. The invertible C-loop codes for two sets of tail fiber genes and provides a means of extending host range.
Species demarcation criteria in the genus
Species differ in host range and tail length (phage P1, 228 nm; phage P1D, 240 nm; and phage 43, 170 nm).
List of species in the genus “P1-like viruses”
| Aeromonas phage 43 |
|
|
| Aeromonas phage 43 |
| (43) |
| Enterobacteria phage P1 |
|
|
| Enterobacteria phage P1 | [AF234172] | (P1) |
| Enterobacteria phage P1D |
| (P1D) |
| Enterobacteria phage P7 |
| (P7) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “P1-like” viruses but have not been approved as species
| Acetobacter phage pKG-2 | (pKG-2) |
| Acetobacter phage pKG-3 | (pKG-3) |
| Enterobacteria phage D6 | (D6) |
| Enterobacteria phage PhiW39 | (PhiW39) |
| Enterobacteria phage j2 | (j2) |
| Pseudomonas phage PP8 | (PP8) |
| Vibrio phage PhiVP25 | (PhiVP253) |
| Vibrio phage P147 | (P147) |
Genus “P2-like viruses”
Type species Enterobacteria phage P2
Distinguishing features
Virion DNA has cohesive ends. Transcription of virion structural genes is divergent.
Virion properties
Morphology
Phage heads are icosahedral, measure about 60 nm in diameter, and consist of 72 capsomers (60 hexamers and 12 pentamers; T=7). Tails measure 135×18nm and have a collar and six short kinked fibers.
Physicochemical and Physical Properties
Virion Mr is 58×106; buoyant density in CsC1 is 1.43 g cm−3; and S20,W is 283S.
Nucleic Acid
Genomes are about 34 kbp, constitute about 48% of particle weight, and have a G+C content of 52%. The genomes of P2 and the related phages (HP1, HP2, 186, φCTX, Fels-2 and K139) have been sequenced.
Proteins
Virions contain at least 13 structural proteins (20–94 kDa), including 420 copies of the major CP (39 kDa). Amino acid sequences of the proteins of phages with completely sequenced genomes are available at GenBank and EMBL.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genome is linear and non-permuted, has cos sites and includes about 40 genes. Transcription starts in the right half of the genome, has two phases (early and late) and depends on host RNA polymerase. Replication starts at a single site, is unidirectional and follows a modified rolling-circle mechanism. DNA is cut from concatemers at specific sites during packaging into proheads.
Antigenic properties
Virions of phages P2, P1 (genus “P1-like viruses”) and Mu (genus “Mu-like viruses”) share tail fiber antigens.
Biological properties
Phages are temperate, adsorb to the cell wall and infect enteric and related Gram-negative bacteria. Prophages may integrate at about 10 specific sites of the bacterial chromosome and are not UV-inducible. P2 acts as a "helper" for defective Enterobacteria phage P4 (P4) by providing head and tail genes for its propagation.
Species demarcation criteria in the genus
Species differ in host range and DNA sequence.
List of species in the genus “P2-like viruses”
| Enterobacteria phage P2 |
|
|
| Enterobacteria phage P2 | [AF063097] | (P2) |
| Haemophilus phage HP1 |
| (HP1) |
| Haemophilus phage HP1 | [U24159] | (HP1) |
| Haemophilus phage S2 |
| (S2) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “P2-like viruses” but have not been approved as species
| Aeromonas phage 29 |
| (29) |
| Aeromonas phage 37 |
| (37) |
| Agrobacterium phage PIIBNV6 |
| (PIIBNV6) |
| Caulobacter phage PhiCr24 |
| (PhiCr24) |
| Enterobacteria phage 186 | [U32222] | (186) |
| Enterobacteria phage 299 |
| (299) |
| Enterobacteria phage Beccles |
| (Beccles) |
| Enterobacteria phage Pk2 |
| (Pk2) |
| Enterobacteria phage W-Phi |
| (W-Phi) |
| Haemophilus phage HP2 | [AY027935] | (HP2) |
| Pasteurella phage AU |
| (AU) |
| Pseudomonas phage PhiCTX | [AB008550] | (PhiCTX) |
| Pseudomonas phage PsP3 |
| (PsP3) |
| Rhizobium phage Phi-gal-1/R |
| (Phi-gal-1/R) |
| Rhizobium phage WT1 |
| (WT1) |
| Salmonella phage Fels-2 |
| (Fels-2) |
| Vibrio phage X29 |
| (X29) |
| Vibrio phage K139 | [AF125163] | (K139) |
Genus “Mu-like viruses”
Type species Enterobacteria phage Mu
Distinguishing features
The viral genome contains two terminal, variable sequences of host DNA. It is able to integrate at virtually any site of the host chromosome and generate a wide range of mutations due to its unique mode of DNA replication (replicative transposition). Integration is required for establishment of lysogeny and for DNA replication during lytic development.
Virion properties
Morphology
Virions have icosahedral heads about 60 nm in diameter, contractile tails about 120×18 nm, a baseplate and six short fibers.
Physicochemical and Physical Properties
Virion buoyant density in CsC1 is 1.49 g cm−3.
Nucleic Acid
The phage Mu genome is about 36–40 kbp, corresponding to about 40% of particle weight, has a G+C content of 50–51% and has been sequenced.
Proteins
Particles have 12 structural proteins (20–76 kDa), including the major coat protein (33 kDa).
Lipids
None reported.
Carbohydrates
None reported
Genome organization and replication
The phage Mu genome is linear and includes 55 genes. Related functions cluster together. The genome is non-permuted and heterogeneous, consisting of 36,717 bp of phage-specific DNA flanked at both ends by 0.5–3 kbp of covalently bound segments of host DNA. It contains an invertible segment of about 3 kbp (the G-loop) that is homologous to the invertible C-segment of Enterobacteria phage P1 (P1) DNA. Infecting DNA undergoes either lytic or lysogenic development. Both modes require (random) integration of phage DNA into host DNA, mediated by a phage-encoded transposase. Transcription starts at the left end of the genome and depends on host RNA polymerase. Replication may start at either end of the genome, is semi-conservative and occurs during transposition into new integration sites. Phage heads package integrated, non-concatemeric phage DNA and adjacent host DNA by an atypical headful mechanism. Progeny phage DNA is cut out of the host DNA 100–200 bp away from a phage-coded pac site.
Antigenic relationships
Enterobacteria phages Mu, D108, P1 and P2 have some common tail fiber antigens.
Biological properties
Viruses are temperate and can carry out generalized transduction. They infect enteric and (possibly) other related gram-negative bacteria. The invertible G-loop codes for two sets of tail fibers which provides a means of extending host range. Prophages are not inducible by irradiation with UV light.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus “Mu-like viruses”
| Enterobacteria phage Mu |
|
|
| Enterobacteria phage D108 |
| (D108) |
| Enterobacteria phage Mu | [AF083977] | (Mu) |
| (Enterobacteria phage Mu-1) |
|
|
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “Mu-like viruses” but have not been approved as species
| Pseudomonas phage B3 | [AF232233] | (B3) |
| Pseudomonas phage B39 |
| (B39) |
| Pseudomonas phage D3112 | [AY394005] | (D3112) |
| Pseudomonas phage PM69 |
| (PM69) |
| Vibrio phage VcA3 |
| (VcA3) |
Genus “SPO1-like viruses”
Type species Bacillus phage SPO1
Note on nomenclature
The “O” in the name SPO1 derives from Osaka, where the phage was isolated. It is therefore properly the letter “O” (oh) and not the numeral “0” (zero). However, in the published literature and earlier versions of this taxonomy, the names “SPO1” and SP01” are used interchangeably to refer to the same virus. As a consequence, database searches for SPO1 should always be done with both forms of the name.
Distinguishing features
Members of this genus are large lytic phages. Heads show conspicuous capsomers. DNA is terminally redundant (but not circularly permuted), contains 5-hydroxymethyluracil and codes for a type A (E. coli Pol I) DNA polymerase.
Virion properties
Morphology
Virions have isometric, icosahedral heads of about 94 nm in diameter with conspicuous capsomers. Contractile tails measure 150×18 nm and have a small collar and a 60 nm-wide baseplate.
Physicochemical and Physical Properties
SPO1 virion Mr is about 180×106; buoyant density in CsC1 is 1.54 g cm−3; and S20,w is 794S.
Nucleic Acid
Genomes are linear, carry about 140–160 kbp and those that have been studied have a G+C content of 42%. Thymine is replaced by 5-hydroxymethyluracil in SPO1 DNA.
Proteins
Virions carry about 53 proteins (16 in the head and 28 in the tail and baseplate). Type A DNA polymerase is encoded in the phage genome.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The genome is linear and may contain as many as 200 genes. Related functions cluster together. The genome has a terminally redundancy of about 12 kbp, but is not circularly permuted. After infection, host syntheses are shut off and replication starts at two SPO1 DNA sites. Phage-encoded sigma factors are used to modify and appropriate host RNA polymerase for phage synthesis.
Biological properties
Phages are virulent and so far have been characterized only from Bacillus and Lactobacillus. Distribution is worldwide.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus “SPO1-like viruses”
| Bacillus phage SPO1 |
|
|
| Bacillus phage SPO1 | [FJ230960] | (SPO1) |
| Bacillus phage SP8 |
| (SP8) |
| Bacillus phage SP82 |
| (SP82) |
Species names are in italic script; names of isolates are in roman script. Sequence accessions [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “SPO1-like viruses” but have not been approved as species
| Bacillus phage AR1 | (AR1) |
| Bacillus phage GS1 | (GS1) |
| Bacillus phage I9 | (I9) |
| Bacillus phage NLP-1 | (NLP-1) |
| Bacillus phage SP5 | (SP5) |
| Bacillus phage SW | (SW) |
| Bacillus phage Phi-e | (Phi-e) |
| Bacillus phage Phi25 | (Phi25) |
| Bacillus phage 2C | (2C) |
| Lactobacillus phage 222a | (222a) |
Genus “PhiH-like viruses”
Type species Halobacterium phage PhiH
Distinguishing features
The host is an archaeon. Phage DNA has a pac site, and is circularly permuted and terminally redundant.
Virion properties
Morphology
Virions have isometric heads 64 nm in diameter, tails of 170×18 nm and short tail fibers.
Physicochemical and Physical Properties
Not known.
Nucleic Acid
Genomes are linear, about 59 kbp in size and have a G+C content of 64%. Cytosine is replaced by 5-methylcytosine.
Proteins
Virions have three major proteins (20, 45 and 70 kDa) and 10 minor components.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
Genomes are partially circularly permuted and about 3% terminally redundant and have a pac site. Halobacterium phage H DNA is markedly variable. All DNAs harbor one or more insertion elements, and also include ordinary deletion and insertion variants. Early transcription is regulated by viral antisense mRNA. Replication results in formation of concatemers. Cutting of concatemers at pac sites is inaccurate and produces DNA molecules with imprecisely defined ends.
Antigenic properties
Not known.
Biological properties
Phages are temperate, specific for halobacteria and require the presence of 3.5 M NaCl. Prophages persist as plasmids and are not UV-inducible.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus “PhiH-like viruses”
| Halobacterium phage phiH |
|
| Halobacterium phage phiH | (PhiH) |
Species names are in italic script; names of isolates are in roman script. Assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “PhiH-like viruses” but have not been approved as species
| Halobacterium phage Hs1 (Hs1) | (Hs1) |
Genus “phiKZ-like viruses”
Type species Pseudomonas phage phiKZ
Distinguishing features
Originally isolated in 1975 in Kazakhstan, phage φKZ represents a genus of unusually large and complex virulent phages specifically infecting Pseudomonas species. Virions are very large and have tails surrounded by fibers, while the phage heads contain an inner body of which the function is not yet known. These phages typically have a broad host spectrum and form a very distant branch in the family Myoviridae. Their genomes (>280 kb) are circularly permuted and terminally redundant.
Virion properties
Morphology
Virions have extraordinarily large icosahedral heads of about 1445 Å in diameter and a 1600 Å long tail (Figure 3), which contracts to half of its original length upon infection. Ultrastructural studies of both the head and tail of φKZ revealed that the major capsid protein of φKZ (gp120) is organized into a surface lattice of hexamers with T=27 triangulation. These are similarly shaped and sized as the hexametric building blocks of bacteriophages T4, 29, P22 and HK97. A complex of several proteins was shown to occupy 11 pentameric vertices of the φKZ capsid.
The tail sheath is assembled around the tail tube and is composed of about 44 rings. Each 36.2 Å-thick ring consists of six gp29 subunits and is rotated by 22° with respect to the previous ring. Analogously, despite the fact that the φKZ tail is much longer, the helical parameters of their contractile sheaths which surround their tail tubes are comparable to the T4 tail. The φKZ baseplate is significantly larger than that of T4 and has a rather flat, hexagonal shape with a diameter of 800 Å and a thickness of 350 Å. Nevertheless, φKZ, like T4, has a cell-puncturing device in the middle of its baseplate, which is likely composed of gp181. Six tail fibers are attached to the φKZ baseplate, each with an approximate length of 500 Å.
Finally, two discs with a radius of 160 Å are present in the neck part of the virus. The φKZ neck, like the neck of T4, may be composed of different proteins whose genes have not yet been identified.
Physicochemical and Physical Properties
Phage 2012-1 virion buoyant density is 1.37 g cm−3. The infectivity is chloroform-resistant and heat-sensitive.
Nucleic Acid
Their circularly permuted genomes (280–316 kb) are packed into a highly condensed series of layers, separated by 24 Å, that follow the contour of the inner wall of the capsid.
Proteins
Virions of different genus members contain between at least 64 (φKZ) and 76 (phage 2012-1) different particle proteins, several of which are subjected to proteolytic processing. Several proteins with unanticipated functions, including an RNA polymerase β subunit, a helicase, a ligase, and an exonuclease, are encoded among the virion-associated genes and were identified as virion proteins.
Genome organization and replication
Phages belonging to the “phiKZ-like viruses” have extraordinarily large genomes. The two fully sequenced members, P. aeruginosa phage φKZ and P. chlororaphis phage 2012-1, carry genomes of 280 and 316 kb, respectively encoding 306 and 461 genes. Comparison between the genomes of 2012-1 and φKZ revealed substantial conservation of the genome plan, and a large region with an especially high rate of gene replacement.
They have a remarkable low GC content (36% for φKZ) and only a minor fraction of the encoded genes exhibit similarity to proteins of known function. Most of these conserved gene products, such as dihydrofolate reductase, ribonucleoside diphosphate reductase, thymidylate synthase, thymidylate kinase and deoxycytidine triphosphate deaminase are involved in nucleotide metabolism. However, no virus-encoded DNA polymerase, DNA replication-associated proteins, or single stranded DNA-binding protein were found based on amino acid homology, and they may therefore be strongly divergent from known homologous proteins. The phiKZ genome also encodes six tRNAs specific for Met (AUG), Asn (AAC), Asp (GAC), Leu (TTA), Thr (ACA) and Pro (CCA). In contrast, only a single tRNA (Leu; anticodon=UAA) is encoded in 2012-1.
Antigenic properties
None reported.
Biological properties
Phages are virulent general transducers and specific for Pseudomonas bacteria, occur in water and soil, and have a worldwide distribution. Although there are only two sequenced members, over 20 similar giant phages with various geographic origins and infecting P. aeruginosa, P. fluorescens, P. chlororaphis and P. stutzeri have been reported. Plaques are clear and very small (0.1 mm in diameter).
Species demarcation criteria in the genus
The identified species share limited or no DNA homology.
List of species in the genus “phiKZ-like viruses”
| Pseudomonas phage phiKZ |
|
|
| Pseudomonas phage phiKZ | [AF399011] | (phiKZ) |
| Pseudomonas phage 201phi2-1 |
|
|
| Pseudomonas phage 201phi2-1 | [EU197055] | (201phi2-1) |
| Pseudomonas phage Lin68 |
|
|
| Pseudomonas phage Lin68 |
| (Lin68) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “phiKZ-like viruses” but have not been approved as species
| Pseudomonas phage EL | [AJ697969] | (EL) |
Genus “I3-like viruses”
Type species Mycobacterium phage I3
Distinguishing features
Mycobacteriophage I3 was isolated in 1970 from a soil sample using Mycobacterium smegmatis strain SN2 as host organism. This phage represents a genus of generally transducing myoviruses which specifically infect mycobacteria. They have an unusual morphology with a characteristic short tail, not resembling any other myoviruses. Their genomes (153–164 kb) are significantly larger than any other mycobacteriophage genome, and are circularly permuted. They form a very distinct branch in the Myoviridae family.
Virion properties
Morphology
The phage head of “I3-like viruses” has regular isometric symmetry with a diameter that varies between 74.7 (Bxz1) and 95.4 (Rizal) nm (Figure 4). As a consequence, the capsid volume fluctuates between 163690 and 340961 nm3. Head capsomers can be seen on the edges of the triangular faces. The tail is distinctly shorter compared to other myoviruses, with an average length of 53.4 nm. A typical hollow tail tube running the length of the tail is apparent after contraction. Other components of the tail are a double collar at the neck of the virus particle and a cup-shaped baseplate.
Physicochemical and physical properties
Like many other mycobacteriophage, phage I3 is sensitive to organic solvents. Maximum inactivation of 6 (LOG10 units) orders was observed with butanol, followed by 3 LOG10 inactivation with methanol. Less than 1 LOG10 inactivation was observed with chloroform, ether and benzene. Starting from these observations, Gope and Gopinathan (1982) demonstrated the presence of lipids in phage I3 comprises 42% DNA, 43% proteins and 15% lipids. Total lipid is composed of 69% phospholipids and 31% neutral lipids. The fatty acid composition of the phage differs markedly from that of its host, both in chain length and the degree of saturation. The phage lipid is mostly composed of saturated fatty acids of which more than 50% are short chain fatty acids.
Nucleic acid
Members of this genus have circularly permuted genomes (153–164 kb). Phage I3 was shown to have modified bases (5-methylcytosine) and 13 to 14 single stranded gaps of about 10 nucleo-tides long.
Proteins
An analysis of the phage protein content is not yet reported.
Lipids
Unconfirmed.
Carbohydrates
None reported.
Genome organization and replication
Phage I3 is the first isolated phage of this genus, but its genome has not yet been fully sequenced. The first sequenced “I3-like virus” phage genome was that of Bxz1, which was recently followed by genomes of six other related phages. Phages of this genus have genomes with GC content varying between 64.7 and 65.4% and which encode between 218 and 229 ORFs. Maximal 8% (Bxz1) of the genome is noncoding sequence.
These phages have very few genes in common with other mycobacteriophages, and only 15% of the ORFs have a predicted function. One of the most striking genomic features is the presence of 26–31 tRNA genes, which is among the highest number identified in any bacteriophage sequenced thus far. The tRNAs in Bxz1 carry anticodons for 15 amino acids. In contrast to phage T4, the genes for tRNAs are not clustered, but are rather scattered in sets of small groups. It was shown that the Bxz1-specific tRNAs modulate the optimal expression of its proteins during development.
Other genomic features include the presence of adenylosuccinate synthase homologs among the Bxz1 subgroup (gp250) and its absence in the genome of Myrna. The latter possesses several proteins not present in the Bxz1 group, including the large hypothetical proteins gp187 (YP_002225066.1) and gp243 (YP_002225120.1), a putative nicotinate phosphoribosyltransferase (gp263, YP_002225140.1) and ATP-dependent protease (gp262, YP_002225139.1).
Bxz1 gp220 encodes a homolog of the human Ro protein, a major target of the autoimmune response in Lupus and Sjögren’s diseases. This suggests that bacteriophages could act in concert with their hosts to stimulate autoimmunity.
Antigenic properties
None reported.
Biological properties
Phages are virulent general transducers and specific for mycobacteria, occur in water and soil and have a worldwide distribution. Phage Bxz1 and its relatives form hazy plaques, although it is unclear whether the cellular survivors are uninfected cells, resistant mutants, or lysogens. Phage I3 was shown to be specific for cell-wall-associated glycopeptidolipid. A single methylated rhamnose was critical for phage binding. The presence of lipids may facilitate the phage–host interaction.
Species demarcation criteria in the genus
Within the “I3-like viruses”, two species can be delineated based on sequence similarity. Phages Cali, Catera, Rizal, ScottMcG show >90% protein similarity to Bxz1 and can be considered as one species (see below). Mycobacteriophage Myrna, with a genome of 164 kb, shares approximately 45% of proteins with the Bxz1 subgroup phages.
List of species in the genus “I3-like viruses”
| Mycobacterium phage I3 |
|
|
| Mycobacterium phage I3 |
| (I3) |
| Mycobacterium phage Bxz1 | [AY129337] | (Bxz1) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus “I3-like viruses” but have not been approved as species
None reported.
List of unassigned species in the family Myoviridae
| Bacillus phage G |
|
|
| Bacillus phage G |
| (G) |
| Bacillus phage PBS1 |
|
|
| Bacillus phage PBS1 |
| (PBS1) |
| Microcystis aeruginosa phage Ma-LMM01 |
|
|
| Microcystis aeruginosa phage Ma-LMM01 | [AB231700] | (Ma-LMM01) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the family Myoviridae but have not been approved as species
| Acinetobacter phage A3/2 |
| (A3/2) |
| Acinetobacter phage A10/45 |
| (A10/45) |
| Acinetobacter phage BS46 |
| (BS46) |
| Acinetobacter phage E14 |
| (E14) |
| Actinomycetes phage SK1 |
| (SK1) |
| Actinomycetes phage 108/016 |
| (108/016) |
| Aeromonas phage Aeh2 |
| (Aeh2) |
| Aeromonas phage 29 |
| (29) |
| Aeromonas phage 51 |
| (51) |
| Aeromonas phage 59.1 |
| (59.1) |
| Alcaligenes phage A6 |
| (A6) |
| Bacillus phage Bace-11 |
| (Bace-11) |
| Bacillus phage CP-54 |
| (CP-54) |
| Bacillus phage MP13 |
| (MP13) |
| Bacillus phage SP3 |
| (SP3) |
| Bacillus phage SP10 |
| (SP10) |
| Bacillus phage SP15 |
| (SP15) |
| Bacillus phage SP50 |
| (SP50) |
| Bacillus phage Spy-2 |
| (Spy-2) |
| Bacillus phage Spy-3 |
| (Spy-3) |
| Bacillus phage SST |
| (SST) |
| Clostridium phage HM3 |
| (HM3) |
| Clostridium phage CE-beta |
| (CE-beta) |
| Coryneform phage A19 |
| (A19) |
| Cyanobacteria phage AS-1 |
| (AS-1) |
| Cyanobacteria phage N1 |
| (N1) |
| Cyanobacteria phage S-6(L) |
| (S-6(L)) |
| Enterobacteria phage FC3-9 |
| (FC3-9) |
| Enterobacteria phage Kl9 |
| (Kl9) |
| Enterobacteria phage PhiP27 | [AJ298298] | (PhiP27) |
| Enterobacteria phage 01 |
| (01) |
| Enterobacteria phage ViI |
| (ViI) |
| Enterobacteria phage Phi92 |
| (Phi92) |
| Enterobacteria phage 121 |
| (121) |
| Enterobacteria phage 16-19 |
| (16-19) |
| Enterobacteria phage 9266 |
| (9266) |
| Halorubrum phage HF2 | [AF222060] | (HF2) |
| Lactobacillus phage fri |
| (fri) |
| Lactobacillus phage hv |
| (hv) |
| Lactobacillus phage hw |
| (hw) |
| Listeria phage A511 |
| (A511) |
| Listeria phage 4211 |
| (4211) |
| Mollicutes phage Br1 |
| (Br1) |
| Pseudomonas phage PB-1 |
| (PB-1) |
| Pseudomonas phage PS17 |
| (PS17) |
| Pseudomonas phage PhiW-14 |
| (PhiW-14) |
| Pseudomonas phage 12S |
| (12S) |
| Rhizobium phage CM1 |
| (CM1) |
| Rhizobium phage CT4 |
| (CT4) |
| Rhizobium phage m |
| (m) |
| Shigella phage SfV | [AF339141] | (SfV) |
| Xanthomonas phage XP5 |
| (XP5) |
| Vibrio phage kappa |
| (kappa) |
| Vibrio phage 06N-22P |
| (06N-22P) |
| Vibrio phage VP1 |
| (VP1) |
| Vibrio phage II |
| (II) |
Phylogenetic relationships within the family
No information available.
Similarity with other taxa
See Order Caudovirales.
Derivation of name
Myo: from Greek my, myos, “muscle”, referring to the contractile tail.
Further reading
Ackermann, H.-W. and DuBow, M.S. (Eds.) (1987). Viruses of Prokaryotes, Vol. II. Natural Groups of Bacteriophages. CRC Press, Boca Raton, FL.
Karam, J.-D. (Ed.) (1994). Molecular Biology of Bacteriophage T4. American Society for Microbiology, Washington, DC.
Klumpp, J., Lavigne, R., Loessner, M.J. and Ackermann, H.W. (2010). The SPO1-related bacteriophages. Arch. Virol., 155, 1547-1561.
Miller, E., Kutter, E., Mosig, G., Arisaka, F., Kunisawa, T. and Rüger, W. (2003). Bacteriophage T4. Microbiol. Molec. Biol. Rev.I 67, 86-156.
Morgan, G., Hatfull, G., Casjens, S. and Hendrix, R. (2002). Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol., 317, 337-359.
Mosig, G. and Eiserling, F. (2006) T4 and related phages: structure and development. In: R. Calendar and S. Abedon (Eds.), The Bacteriophages, 2nd edn. Oxford University Press, New York, pp. 225-268.
Nilsson, A.S. and Ljungquist, E.H. (2006) The P2-like bacteriophages. In: R. Calendar and S. Abedon (Eds.), The Bacteriophages, 2nd edn. Oxford University Press, New York, pp. 365-391.
Paolozzi, L. and Ghelardini, P. (2006) The bacteriophage Mu. In: R. Calendar and S. Abedon (Eds.), The Bacteriophages, 2nd edn. Oxford University Press, New York, pp. 469-498.
Contributed by
Lavigne, R. and Ceyssens, P-J.
The authors acknowledge the contribution to the Eighth ICTV Report of Hendrix, R.W. and Casjens, S.R.
Figures
Figures
Figure 1 (Left) Diagram of Enterobacteria phage T4 (T4) showing detailed location of structural proteins. Head vertices consist of cleaved gp24. Gp20 is located at the headtail connector. Collar and whiskers appear to be made of the same protein, gpwac. Sheath subunits (gp18) fit into holes in the base plate and short tail proteins (gp12) are shown in the quiescent state. The complex base is assembled from a central plug and six wedges. Tail fibers consist of three proteins. (Right) Negative contrast electron micrograph of T4 particle stained with uranyl acetate. The bars represent 100 nm.
(From Eiserling, F.A. (1983). In: Bacteriophage T4 (C.K. Mathews, E.M. Kutter, G. Mosig and P.B. Berget, Eds.), American Society for Microbiology, Washington, DC; with permission.)
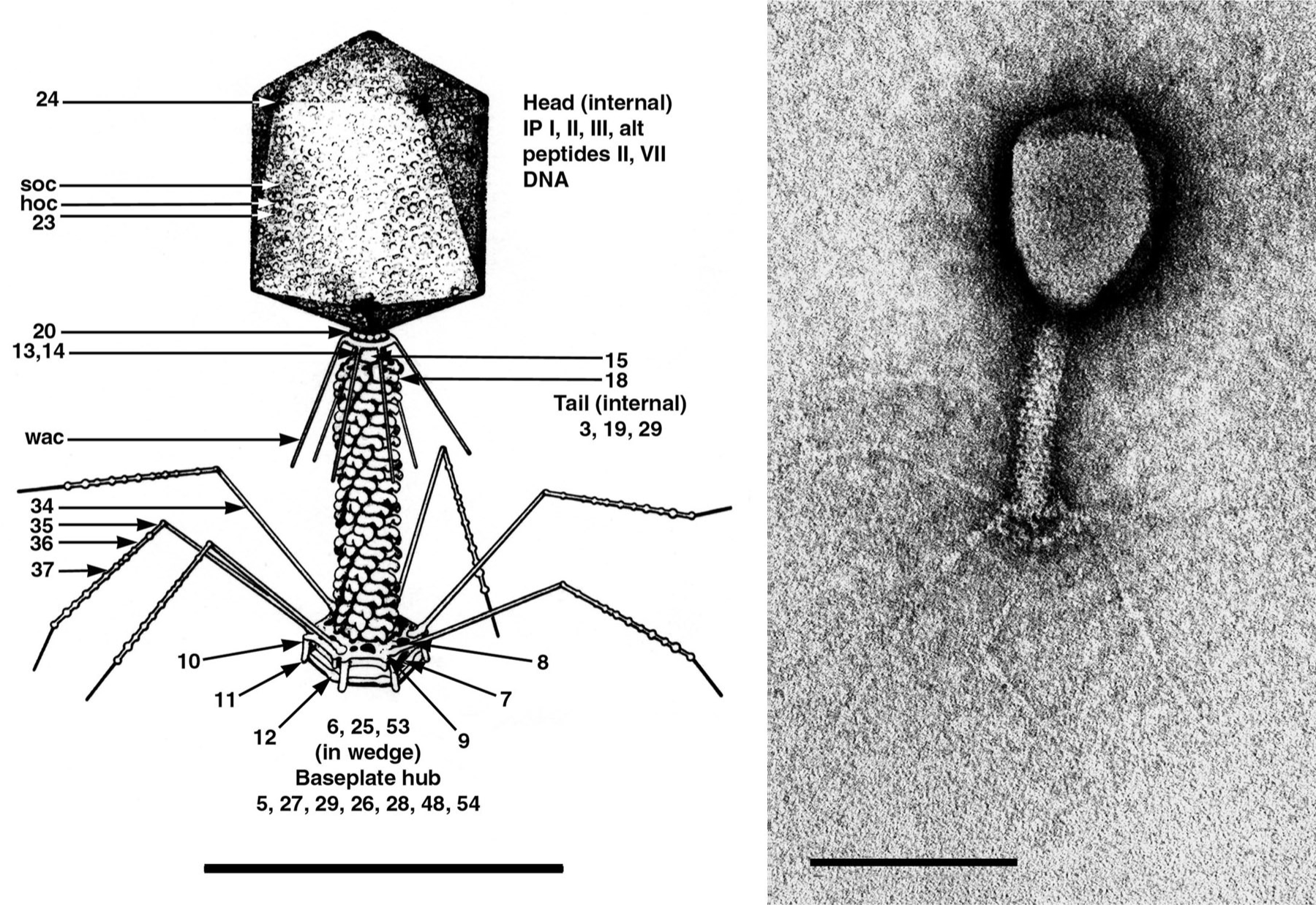
Figure 2 Simplified genetic map of Enterobacteria phage T4 (T4) showing clustering of genes with related functions, location of essential genes (solid bars) and direction and origin of transcripts (arrows).
(From Freifelder, D. (Ed.) (1983). Molecular Biology, Science Books International, Boston, MA, and Van Nostrand Reinhold, New York, p. 614; with permission.)
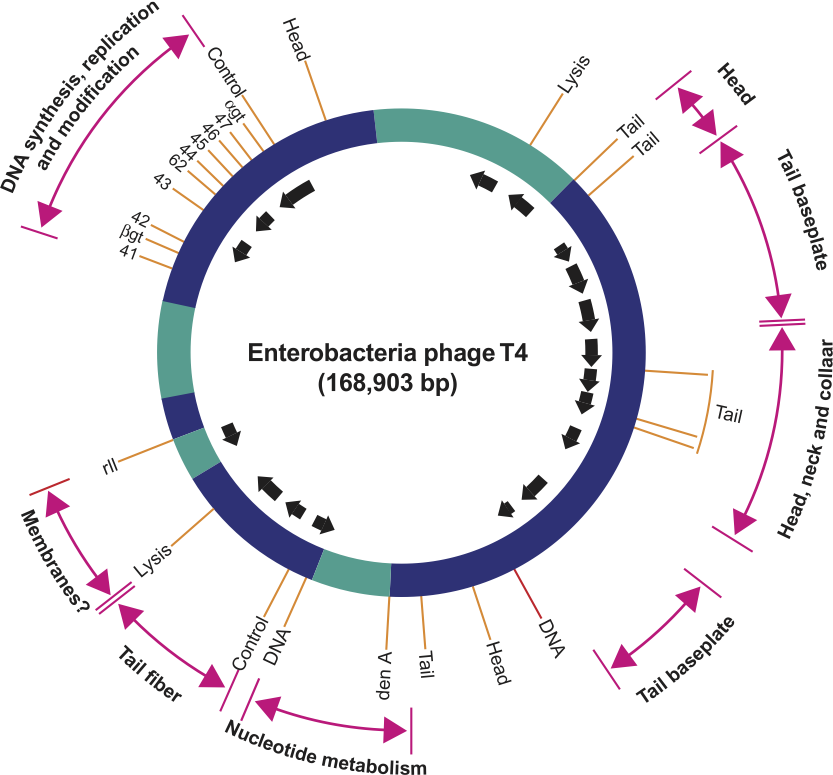
Figure 3 Cryo-EM-based reconstruction of bacteriophage KZ. Spiralling tail sheath vertices are highlighted.
(Reconstruction courtesy of Andrei Fokine.)
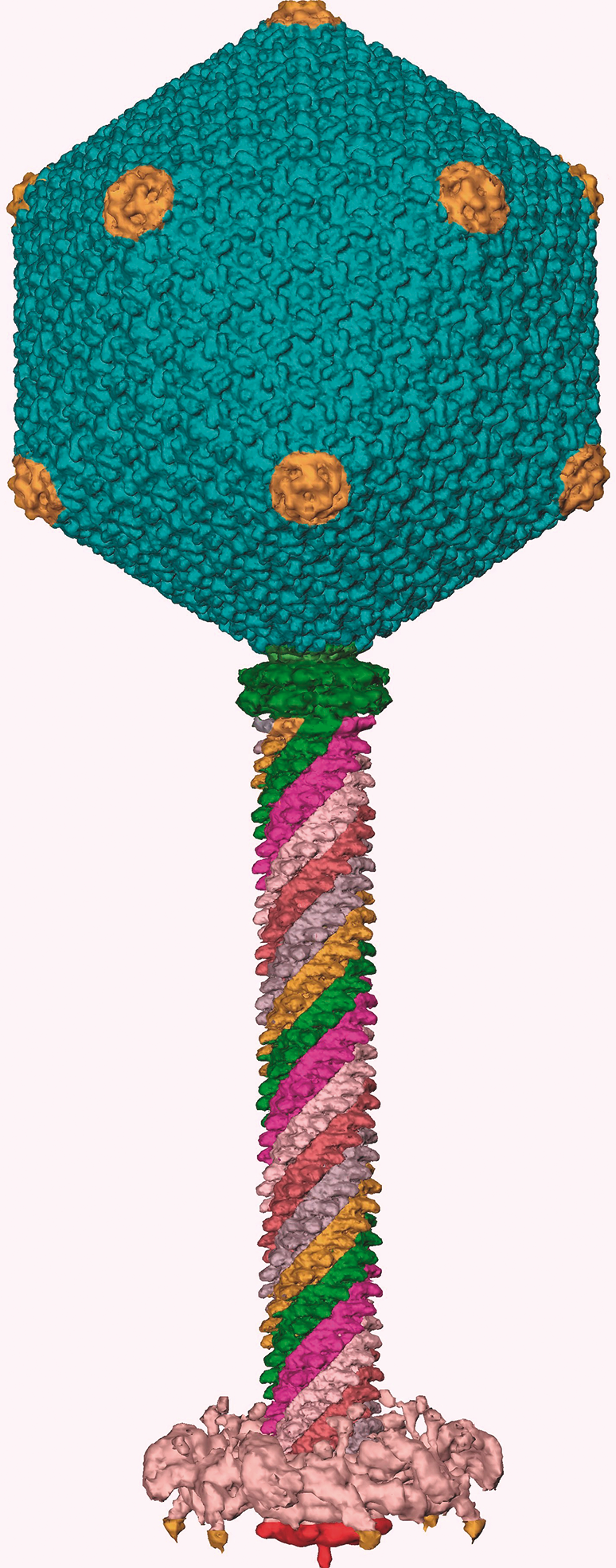
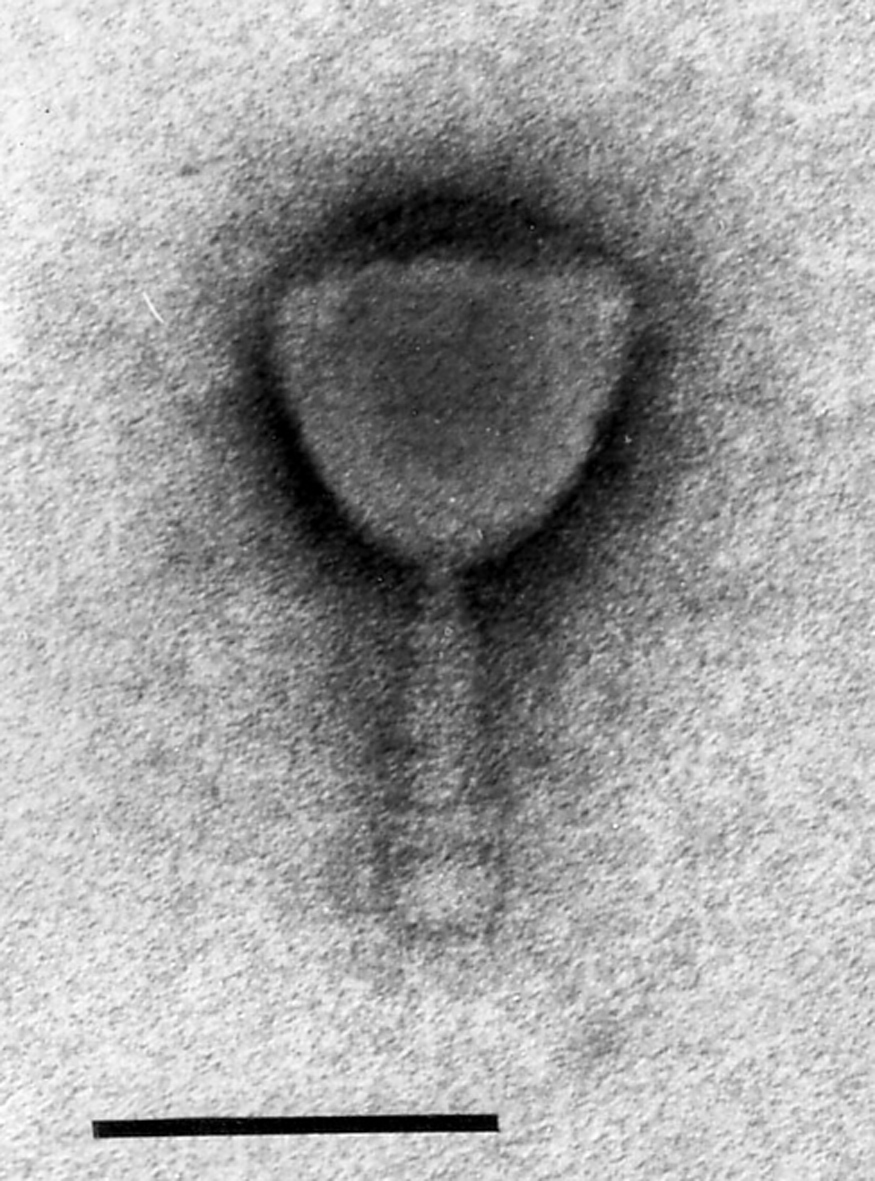
Figure 4 Electron micrographs of Mycobacterium phage I3. Phage particles were positively stained with uranyl acetate. The bar represents 100 nm.
(Image courtesy of H. Ackermann.)