Family: Mimiviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Mimivirus
Type species Acanthamoeba polyphaga mimivirus
Virion properties
Morphology
Mimivirus particles are comprised of an icosahedral core protein capsid with a diameter of 0.5 μm. This large capsid is uniformly covered with a 0.125 μm thick layer of closely packed fibers, forming a roughly spherical object 0.75 μm in diameter that is easily visible under the light microscope with Nomarski optics (differential interference contrast microscopy). The mimivirus particle is not entirely symmetrical as it has a five-fold star-shaped structure at a single icosahedral vertex. This structure, coined the stargate, extends along the whole length of the five icosahedral edges surrounding this unique vertex (Figure 1). The central electron-dense nucleocapsid of the particle appears to be contained within two 40 Å-thick lipid membranes surrounded by a 70 Å-thick protein shell (Figure 2).
Physicochemical and physical properties
Using ultracentrifugation in a CsCI gradient, the particle density was estimated to be about 1.36 g cm−3. In addition to their unprecedented size and hairy appearance, mimivirus particles are highly resistant to extreme conditions (pH, temperature, reducing agents) or enzymatic treatments (proteases, glycosidases). The resilience of the particles may be due to their distinctive peripheral fiber layer thought to be made of a dense mesh of a complex (lipo-) polysaccharide biopolymer akin to the outer coat of bacterial spores. The presence of this outer layer, not seen in other families of large nucleocytoplasmic DNA viruses, together with the overall size of the particle, might be linked to the heterotrophic nature of the host acanthamoeba; mimivirus particles mimic the bacteria on which they usually feed (Figure 2).
Nucleic acid
The mimivirus genome is a single linear dsDNA molecule of 1,181,549 bp with an overall nucleotide composition of 72% (A+T). Several virus-encoded mRNAs are associated with the purified particle, but the functional significance of these findings is unknown. Mimivirus genome termini lack the large terminal inverted repeats (up to 2 kb) found in other large DNA viruses such as phycodnaviruses (chlorovirus) or poxviruses. In place of inverted terminal repeats, the mimivirus genome has an inverted repeat of a 617-bp segment approximately 22 kb from one end of its genome and near the other end. Pairing these regions can lead to a putative Q-like form for the mimivirus genome, with a long (22,514 bp) and a short (259 bp) tail. The short tail does not contain any ORFs.
Proteins
A comprehensive proteomic study of purified virions led to the identification of the products of at least 137 mimivirus genes. Over half of these virion-associated proteins have unknown functions. The 2D gels revealed numerous isoforms, probably due to posttranslational modifications such as glycosylation, acetylation and phosphorylation. In addition to the expected major structural components (e.g., the major capsid protein L425 and core L410 protein), transcription enzymes and factors (12 gene products) constitute the largest functional category associated with the viral particles. This set includes all five predicted DNA-directed RNA polymerase subunits, two helicases (R350, L540), the mRNA capping enzyme and four transcription factors (L377, L538, L544, R563), including a TATA box-like binding protein. The presence of a complete transcription machinery in mimivirus particles is reminiscent of poxviruses. The next largest functional group contains nine gene products associated with oxidative pathways. Protein/lipid modification enzymes are also well represented, including a phosphoesterase and a lipase, which are eventually used for digesting the cell (vacuole) membrane, two protein kinases and a protein phosphatase. Finally, seven proteins associated with DNA topology, damage repair and replication are in the virion, including topoisomerases IA and IB (R194, L221), a DNA UV damage repair endonuclease (L687) and two types (X and B) of DNA polymerases (L318, R322).
Lipids
The interior of the particle appears to exhibit two lipid membranes, one encircling a dark core (nucleic acid) and one in direct contact with the interior of the protein capsid. However, this membrane organization might also result from the tight folding of a single membrane. Once the infecting virion is located within a phagocytic vacuole, the particle stargate opens allowing fusion of at least one of the mimivirus lipid membranes with the vacuole membrane. This fusion directly delivers the core of the particle into the host cytoplasm where it serves as a seed to initiate the formation of a virion factory. As expected from the crucial role likely to be played by the internal lipid membranes of the particle in the infection process, mimivirus virions are readily inactivated by treatment with lipophilic compounds (such as DMSO).
Carbohydrates
The ultra-structure of the mature particle (as observed by electron or atomic-force microscopy) as well as preliminary biochemical and mass spectrometry analyses suggest that the outer fiber layer might consist of a biopolymer similar to peptidoglycan. Accordingly, the mimivirus genome encodes a number of proteins homologous to enzymes key to the synthesis of bacterial cell walls. Among those proteins, the functions of a UDP-D-glucose 4,6-dehydratase (R141) and of a bifunctional UDP-4-keto-6-deoxy-D-glucose epimerase/reductase (L780) have been experimentally validated.
Genome organization and replication
The initial mimivirus genome annotation predicted 911 protein-coding genes and 6 tRNAs (Figure 3). More recent data obtained through transcriptome sequencing (RNA-Seq) and deep genome resequencing allowed the identification of a total of 1018 genes, including 979 protein-coding genes, 6tRNAs and 33 non-coding mRNAs. The latest genome sequence and the most current annotation (including the location of identified promoter signals and known 5′-end and 3′-end transcript boundaries) is available in the RefSeq database under accession number NC_014649.1, and in GenBank under accession number HQ336222.
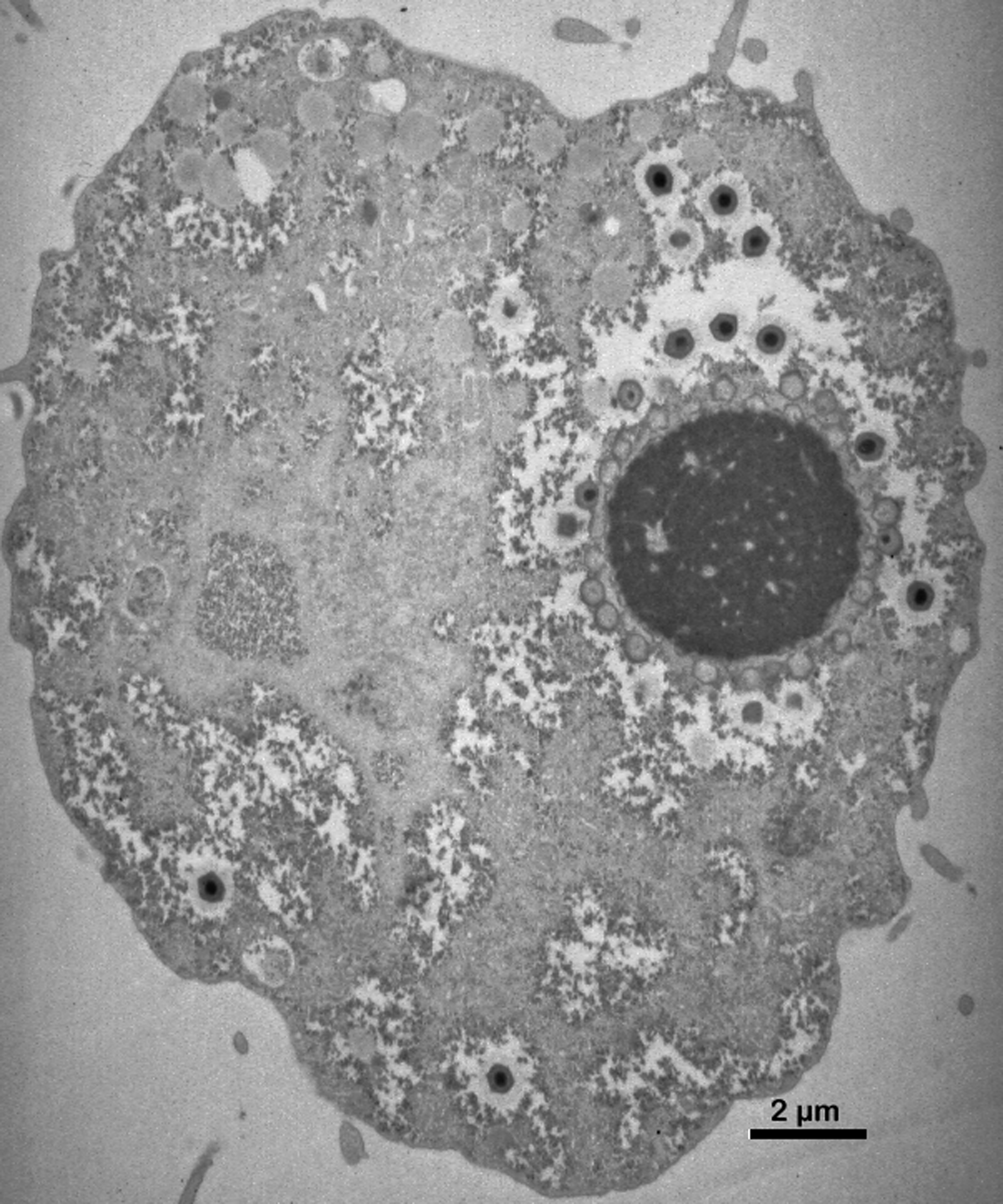
The penetration of the particle inner core within the host cytoplasm is followed by a complete eclipse phase that lasts approximately two hours in Acanthamoeba castellanii (ATCC 30010), after which time mimivirus virion factories become visible. Mimivirus replication entirely takes place in the cytoplasm of the host Acanthamoeba cell, through the successive expression of early (from 0 to 3 h post infection), intermediate (from 3 h to 6 h post-infection) and late (after 6 h post-infection) transcripts, each gene class representing approximately one-third of the mimivirus genome. The virion factories develop from the core of individual uncoated virus particles (seeds). The earliest viral transcripts are detected as soon as 15 minutes post infection, most likely produced by the viral transcription machinery within the uncoated particles. Most of the genes involved in nucleotide synthesis and DNA replication are transcribed from 3 h to 6 h post-infection. Late genes (after 6 h) include virion structural components, as well as most of the virally-encoded transcription apparatus components. This expression pattern suggests that the early and intermediate mimivirus transcripts detected before the appearance of fully mature cytoplasmic virion factories are generated by the transcription apparatus associated with the virion core. Mimivirus particles (at least one thousand per infected cell) are continually produced for up to 12 h by the growing virion factories (up to 6 µm in diameter) (Figure 4). Mature mimivirus particles increasingly fill the host cytoplasm and are progressively released from the dying cell. No budding or sudden cell bursts are seen.
Antigenic properties and pathogenicity
Acanthamoeba polyphaga mimivirus was isolated from the water of a cooling tower in Bradford, England. Mimivirus readily infects many Acanthamoeba strains, including its preferred laboratory host Acanthamoeba castellanii. Metagenomic surveys indicate that close relatives of the Mimiviridae family are prevalent in the sea, where they probably infect marine heterotrophic protists and regulate plankton populations.
Although mimivirus was isolated in the context of a pneumonia epidemic and initially thought to be an emerging human pathogen based on positive serology, subsequent more specific PCR-based studies failed to detect mimivirus in large numbers of pneumonia patients. As expected from the size of its particle, mimivirus is internalized by various professional phagocytic cells, including human macrophages in vitro. However, the recent demonstration that mimivirus particles are recognized by the sera of patients infected by Francisella tularensis, casts serious doubt on previously published serological evidence for mimivirus infection of humans. At the moment, there is little evidence that mimivirus is a human pathogen.
Biological properties
The presence of many proteins never seen in any other virus is one of the unique features of mimivirus. In addition to the full DNA replication and transcription apparatus usually found in large eukaryotic DNA viruses (such as the poxviruses), mimivirus encodes many enzymes central to the translation apparatus such as four aminoacyl-tRNA synthase: Tyr-RS, Met-RS, Arg-RS and Cys-RS. Mimivirus also possesses the first virally-encoded nucleotide diphosphate kinase (NDK), mismatch repair MutS-homolog, and tRNA (Uracil-5-)-methyltransferase. The mimivirus genome also encodes several sugar-manipulating enzymes likely to be involved in the biosynthesis of the particle outer layer. Crystallographic 3D structures have been obtained for the following mimivirus proteins: Tyr-RS (L124), NDK (R418), formamidopyrimidine-DNA glycosylase (L315), sulfhydryl oxidase (R596), mRNA-capping enzyme (R382), and mimivirus cyclophilin (L605). A new type of satellite virus (called the Sputnik virophage) has been isolated in association with a new strain of mimivirus (called mamavirus).
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Mimivirus
| Acanthamoeba polyphaga mimivirus |
|
|
| Acanthamoeba polyphaga mimivirus (Rowbotham-Bradford) | [HQ336222=NC_014649] | (APMV) |
| Acanthamoeba polyphaga mamavirus (La Scola-Paris) | [EU827539, EU827540, EU827541] | (APMV2) |
| Megavirus chilensis |
|
|
| Megavirus chilensis (Claverie-Las Cruces) | [JN258408] | (McV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Mimivirus but have not been approved as species
TerravirusCourdovirusMoumouvirus
Derivation of name
Mimi: for mimicking microbe.
Further reading
Journals and books
Abergel, C., Rudinger-Thirion, J., Giegé, R. and Claverie, J.M. (2007). Virus-encoded aminoacyl-tRNA synthetases: structural and functional characterization of mimivirus TyrRS and MetRS. J. Virol., 81, 12406-12417.
Claverie, J.M. and Abergel, C. (2009). Mimivirus and its virophage. Annu. Rev. Genet., 43, 49-66.
La Scola, B., Desnues, C., Pagnier, I., Robert, C., Barrassi, L., Fournous, G., Merchat, M., Suzan-Monti, M., Forterre, P., Koonin, E. and Raoult, D. (2008). The virophage as a unique parasite of the giant Mimivirus. Nature, 455, 100-104.
Legendre, M., Audic, S., Poirot, O., Byrne, D., Lartigue, A., Lescot, M., Bernadac, A., Poulain, J., Abergel, C. and Claverie, J.M. (2010). mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. Genome Res., 20, 664-674.
Monier, A., Claverie, J.M. and Ogata, H. (2008). Taxonomic distribution of large DNA viruses in the sea. Genome Biol., 9, R106.
Mutsafi, Y., Zauberman, N., Sabanay, I. and Minsky, A. (2010). Vaccinia-like cytoplasmic replication of the giant Mimivirus. Proc. Natl Acad. Sci., U S A, 107, 5978-5982.
Pelletier, N., Raoult, D. and La Scola, B. (2009). Specific recognition of the major capsid protein of Acanthamoeba polyphaga Mimivirus by sera of patients infected by Francisella tularensis. FEMS Microbiol Lett., 297, 117-123.
Raoult, D., Audic, S., Robert, C., Abergel, C., Renesto, P., Ogata, H., La Scola, B., Suzan, M. and Claverie, J.M. (2004). The 1.2-Mb genome sequence of mimivirus. Science, 306, 1344-1350.
Xiao, C, Kuznetsov, YG, Sun, S, Hafenstein, S.L., Kostyuchenko, V.A., Chipman, P.R., Suzan-Monti, M., Raoult, D., McPherson, A. and Rossmann, M.G. (2009). Structural studies of the giant mimivirus. PLoS Biol., 7, e92.
Zauberman, N., Mutsafi, Y., Halevy, D.B., Shimoni, E., Klein, E., Xiao, C., Sun, S. and Minsky, A. (2008). Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol., 6, e114.
Websites
Results for Mimivirus at http://www.igs.cnrs-mrs.fr
Contributed by
Claverie, J-M. and Abergel, C.
Figures
Figure 1 Mimivirus particle exhibiting the distinctive stargate structure (transmission electron microscopy).
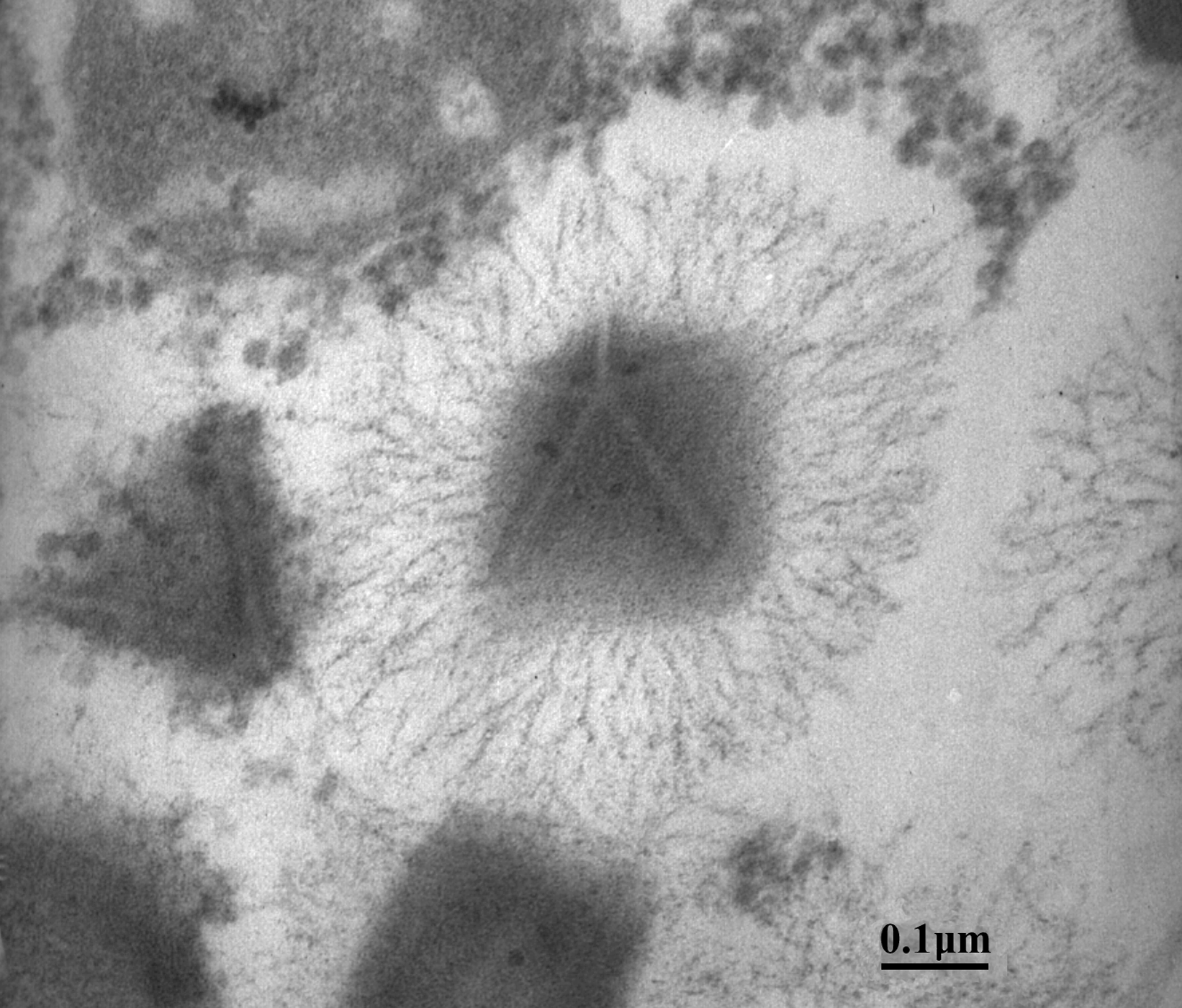
Figure 2 The complex internal structure of a mimivirus particle (transmission electron microscopy).
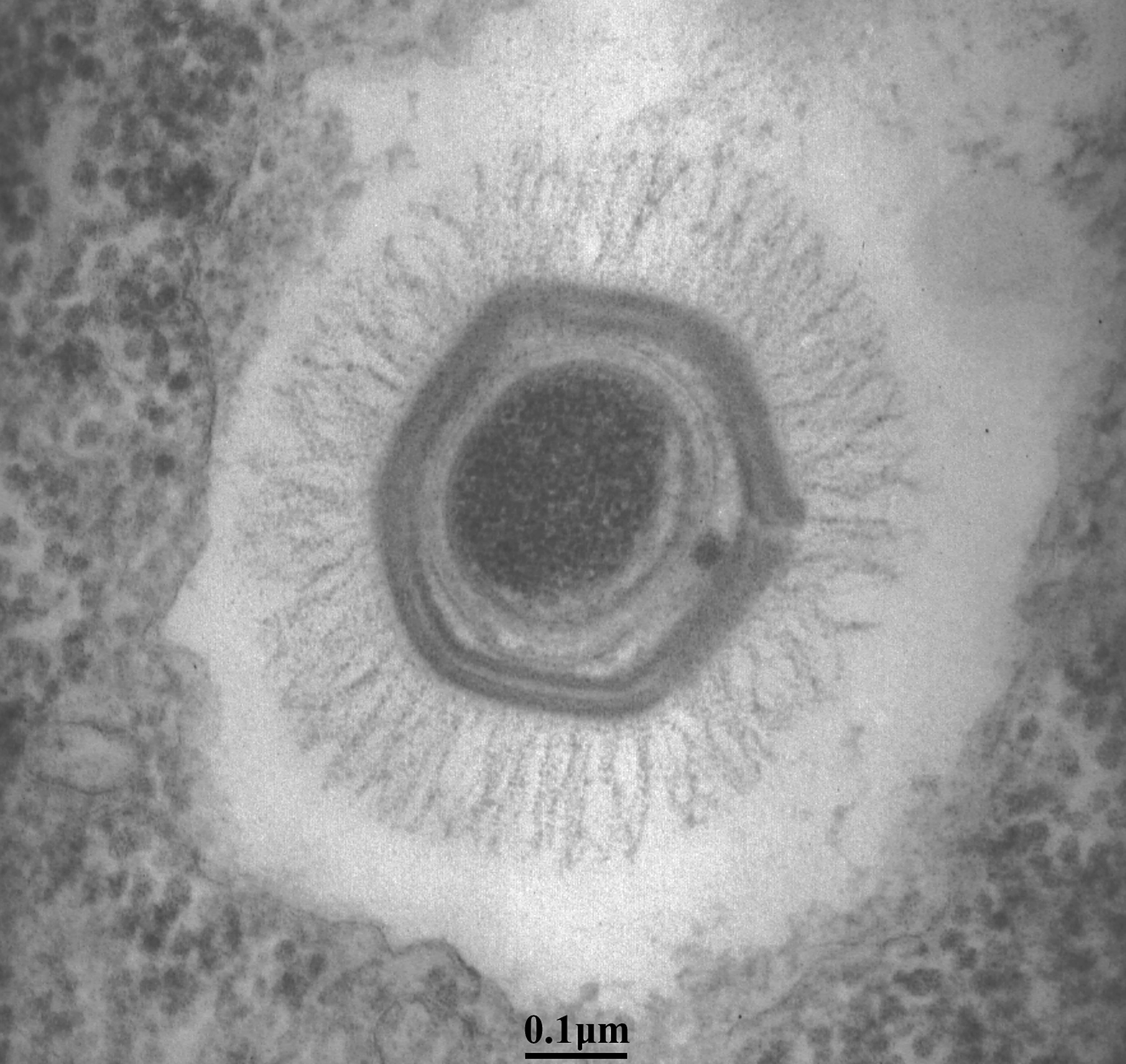
Figure 3 Map of the mimivirus chromosome. The predicted protein coding sequences are shown on both strands and colored according to the function category of their matching COG. Genes with no COG match are shown in gray. Abbreviations for the COG functional categories are as follows: E, amino acid transport and metabolism; F, nucleotide transport and metabolism; J, translation; K, transcription; L, replication, recombination, and repair; M, cell wall/membrane biogenesis; N, cell motility; O, posttranslational modification, protein turnover, and chaperones; Q, secondary metabolites biosynthesis, transport, and catabolism; R, general function prediction only; S, function unknown. Small red arrows indicate the location and orientation of tRNAs. The A+C excess profile is shown on the innermost circle, exhibiting a peak around position 380,000.
(From Raoult et al. (2004). Science, 306, 13441350; with permission of AAAS.)
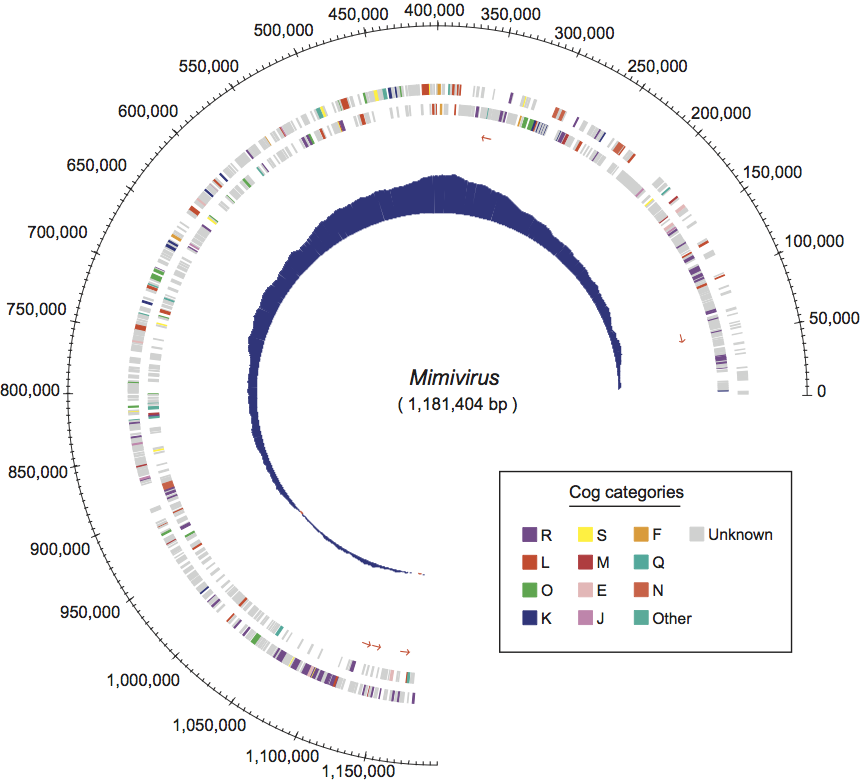
Figure 4 The distinctive giant mimivirus virion factory in full production (8 h post infection in Acanthamoeba castellanii). The dark circle (about 4.5 m in diameter) is the virion factory from which mimivirus particles can be seen emerging, first empty, then filled with a dense core, then covered with their outer fiber layer (transmission electron microscopy).