Family: Iridoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Iridovirus particles consist, from inside out, of an inner DNA/protein core, an internal limiting membrane, a viral capsid, and, in the case of those particles that bud from the plasma membrane, an outer viral envelope (Figure 1). Virions display icosahedral symmetry and are usually 120–200 nm in diameter, but may be up to 350 nm (e.g. genus Lymphocystivirus). The vitrified virion of invertebrate iridescent virus 6 (IIV-6) has diameters of 162, 165 and 185 nm along the two-, three- and five-fold axes of symmetry, respectively (Figure 2A, B). The virion core is an electron-dense entity consisting of a nucleoprotein filament surrounded by a lipid membrane containing transmembrane proteins of unknown function (Figure 1; Figure 2). Each particle is formed of 12 pentasymmetrons and 20 trisymmetrons arranged in an icosahedral, quasi-equivalent symmetry with triangulation number T=147 (h=7, k=7). Both types of structure predominantly comprise hexavalent capsomers, a total of 1460 per virion, that are composed of the major capsid protein (MCP). Each hexavalent capsomer is formed by a non-covalent MCP trimer on the outer surface and a second MCP trimer linked by disulfide bonds on the inner surface. Each MCP monomer contains two beta-barrel domains of the viral jelly-roll type. The 55 capsomers in a trisymmetron are uniformly packed with an intercapsomer distance of 7.5 nm and in a single shared orientation, which is rotated by about 60° compared to the capsomers of the neighbouring trisymmetron. In addition, each pentasymmetron comprises 30 hexavalent (trimeric) capsomers and a single pentavalent capsomer at its centre, at the vertex of each pentasymmetron, for a total of 12 in each virion. The pentavalent capsomer is significantly larger than the trimeric capsomers and has a five-bladed propeller-shaped external appearance and a small central pore that opens into a flask-shaped cavity (Figure 2D, E, F).
A number of additional proteins have recently been identified in the capsid shell and in association with the lipid membrane (Figure 3). These have been named zip monomers, zip dimers, finger proteins and anchor proteins. The zip dimers appear as two halves of a clasp connecting the trimer capsomers along the edges of adjacent trisymmetrons, whereas zip monomers appear to be involved in linking trisymmetron capsomers with those of neighbouring pentasymmetrons. Three sets of nine inward-pointing finger proteins bind the capsomers along the edges of each trisymmetron. Finally, an anchor protein connects each pentasymmetron with the lipid membrane at a distance of two capsomers from the pentavalent vertex (Figure 2G; Figure 3). Other transmembrane proteins are present, but only the anchor protein can be visualized in image reconstruction studies due to its invariable position with respect to the symmetry of the particle. Volume estimates using the MCP as a reference suggest molecular masses of 11.9, 19.7 and 32.4 kDa for the zip, finger and anchor proteins, respectively.
The outer surface of the capsid is covered by flexible fibrils or fibres. Conventional EM studies have suggested that these fibrils are often rather short (ca. 2.5 nm in length) and may have terminal knobs, but in IIV-6 a single fibril of 2 nm width and about 35 nm in length extends outwards from the centre of each trimeric capsomer (Figure 2F; Figure 3). The role of the fibrils remains unknown.
Iridovirids (a generic term describing all members of the family) may acquire an envelope by budding through the host cell membrane. The envelope increases the specific infectivity of virions, but is not required for infectivity as naked particles are also infectious.
Physicochemical and physical properties
The Mr of virions is 1.05–2.75×109, their sedimentation coefficient (S20,w) is 2020–4460S, and their density is 1.26–1.60 g cm−3. Virions are stable in water at 4 °C for extended periods. Sensitivity to pH varies, whereas sensitivity to ether and chloroform depends on the assay system employed. All viruses are inactivated within 30 min at >55 °C. Frog virus 3 (FV-3), infectious spleen and kidney necrosis virus (ISKNV), invertebrate iridescent virus 6 (IIV-6) and likely other members of the family are inactivated by UV-irradiation. Some ranaviruses remain infectious after desiccation, e.g., Bohle iridovirus (BIV) survives desiccation at temperatures up to 42 °C for up to 6 weeks, whereas others are sensitive to drying.
Nucleic acid
The virion core contains a single linear dsDNA molecule of 140–303 kbp, a value that includes both unique and terminally redundant sequences. However, when considering only the non-redundant portion, genome sizes range from 105 to 212 kbp (Table 1). An isolate of invertebrate iridescent virus 1 (IIV-1) has been reported to have an additional genetic component of 10.8 kbp which exists as a free molecule in the particle core. DNA comprises 12–16% of the particle weight, and the G+C content ranges from about 28 to about 55%. All viruses within the family possess genomes that are circularly permuted and terminally redundant. However, the DNA of vertebrate iridoviruses is highly methylated, whereas little to no methylation is found within the genomes of the invertebrate iridoviruses. The complete genomic sequence is known for 15 iridoviruses, with representative sequence information available from every genus in the family Iridoviridae. The iridoviruses whose genomes have been completely sequenced include IIV-6, IIV-3, lymphocystis disease virus 1 (LCDV-1), LCDV-China (LCDV-C), ISKNV, orange spotted grouper iridovirus (OSGIV), rock bream iridovirus (RBIV), red seabream iridovirus (RSIV), grouper iridovirus (GIV), Singapore-GIV (SGIV), tiger frog virus (TFV), frog virus 3 (FV-3), epizootic hematopoietic necrosis virus (EHNV), soft-shelled turtle iridovirus (STIV) and Ambystoma tigrinum virus (ATV). Although naked genomic DNA is not infectious, non-genetic reactivation of viral DNA can be achieved in the presence of viral structural proteins.
Table 1 Summary of completely sequenced iridovirid genomes
| Genus/Virus | Genome size (kb) | GC content (%) | Number of potential genes | Accession No. |
| Ranavirus |
|
|
|
|
| ATV | 106,332 | 54 | 92 | AY150217 |
| EHNV | 127,011 | 54 | 100 | FJ433873 |
| FV-3 | 105,903 | 55 | 97 | AY548484 |
| TFV | 105,057 | 55 | 103 | AF389451 |
| STIV | 105,890 | 55 | 105 | EU627010 |
| SGIV | 140,131 | 48 | 139 | AY521625 |
| GIV | 139,793 | 49 | 139 | AY666015 |
| Megalocytivirus |
|
|
|
|
| ISKNV | 111,362 | 55 | 117 | AF371960 |
| OSGIV | 112,636 | 54 | 116 | AY894343 |
| RBIV | 112,080 | 53 | 116 | AY532606 |
| RSIV | 112,415 | 53 | 116 | BD143114 |
| Lymphocystivirus |
|
|
|
|
| LCDV-1 | 102,653 | 29 | 108 | L63545 |
| LCDV-C | 186,247 | 27 | 178 | AY380826 |
| Iridovirus |
|
|
|
|
| IIV-6 | 212,482 | 29 | 211 | AF303741 |
| Chloriridovirus |
|
|
|
|
| IIV-3 | 190,132 | 48 | 126 | DQ643392 |
For definitions of abbreviations, please refer to lists of species names, below.
Proteins
Iridoviruses are structurally complex, and up to 36 polypeptides, ranging from about 5 to 250 kDa, have been detected by two-dimensional PAGE of virus particles. Sequence analysis has identified more than 100 ORFs (Tables 1 and 2, Figure 4). The major capsid protein (MCP), 48–55 kDa, comprises 40% of the total virion protein, and its complete amino acid (aa) sequence is known for several viruses. The MCP is highly conserved and shares aa sequence identity with the MCPs of African swine fever virus (ASFV, family Asfarviridae), several members of the family Ascoviridae, and Paramecium bursaria Chlorella virus 1 (PBCV-1, family Phycodnaviridae). At least six DNA-associated polypeptides have been identified in the core of IIV-6, with a major species of 12.5 kDa. A virion-associated protein elicits the shutdown of host macromolecular synthesis, whereas other virion-associated proteins transactivate FV-3 early viral transcription. A number of virion-associated enzymatic activities have been detected, including a protein kinase, nucleotide phosphohydrolase, a ss/dsRNA-specific ribonuclease, pH 5 and pH 7.5 deoxyribonucleases, and a protein phosphatase. In addition to these polypeptides, various other proteins have been identified by BLAST analysis of recently sequenced viral genomes (Table 2; Figure 4).
Table 2 Partial listing of putative gene products encoded by viruses within the genera Iridovirus, Chloriridovirus, Ranavirus, Lymphocystivirus and Megalocytivirus
| Category | Gene product | ||||||
|
| IIV-6 | IIV-3 | ATV | TFV | LCDV-1 | LCDV-C | ISKNV |
| Enzymes associated with nucleic acid replication and metabolism |
|
|
|
|
|
|
|
| DNA polymerase | + | + | + | + | + | + | + |
| RNA polymerase II, α subunit | + | + | + | + | + | + | + |
| RNA polymerase II, β subunit | + | + | + | + | + | + | + |
| Transcription factor-like protein | + | + | + | + | + | + |
|
| RAD-2, DNA repair enzyme | + | + | + | + | + | + | + |
| Cytosine DNA methyltransferase |
|
| + | + | + | + | + |
| Type II restriction enzyme Msp I of Moraxella sp. |
|
|
|
| + |
|
|
| RNAse III | + | + | + | + | + | + | + |
| Ribonucleotide reductase, large subunit | + | + | + | + | + | + |
|
| Ribonucleotide reductase, small subunit | + | + | + | + | + | + | + |
| DUTPase | + |
| + | + |
|
|
|
| Thymidylate synthase | + |
| + | + |
| + |
|
| Thymidine kinase | + | + |
|
| + |
|
|
| Thymidylate kinase | + |
| + |
|
|
|
|
| Topoisomerase II-like protein | + | + |
|
|
|
|
|
| Helicase | + | + | + | + |
| + |
|
| PCNA protein | + | + | + | + | + | + | + |
| DNA ligase | + | + |
|
|
|
|
|
| Additional enzymatic activities |
|
|
|
|
|
|
|
| Tyrosine phosphatase | + |
|
|
|
|
|
|
| Tyrosine kinase | + | + | + | + | + | + | + |
| Thiol oxidoreductase | + | + | + | + | + | + | + |
| Serine/threonine protein kinase | + | + | + | + | + | + | + |
| ATPase | + |
| + | + | + | + | + |
| Matrix metalloproteinase | + | + |
|
|
|
|
|
| mRNA capping enzyme |
|
|
|
|
|
| + |
| Cathepsin B-like protein | + |
|
|
| + |
|
|
| Acetyl-coenyme A hydrolase |
|
|
|
|
| + |
|
| Putative immune evasion proteins |
|
|
|
|
|
|
|
| TNF receptor-associated factor |
|
|
|
|
| + | + |
| Growth factor/cytokine receptor family signature | + |
|
|
|
|
|
|
| TNFR/NGFR family proteins |
|
|
|
| + |
|
|
| PDGF/VEGF-like protein |
|
|
|
|
|
| + |
| Apoptosis inhibitor (IAP) of Cydia pomonellla granulosis virus, Bir repeat profile | + |
|
|
|
|
|
|
| IAP-like protein of African swine fever virus | + |
|
|
|
|
|
|
| CARD, caspase recruitment domain |
|
| + |
|
| + |
|
| 3β-hydroxy-Δ5-C27-steroid oxidoreductase |
|
| + | + | + | + |
|
| eIF-2 α homolog |
|
| + | + |
|
|
|
| Src homology domain, suppressor of cytokine signaling |
|
|
|
|
|
| + |
| Structural proteins |
|
|
|
|
|
|
|
| Major capsid protein | + | + | + | + | + | + | + |
| Myristylated membrane protein | + | + | + | + | + | + | + |
The presence of a homolog of the indicated gene is indicated by a plus sign (+).
Lipids
Non-enveloped particles contain 5–17% lipid, predominantly as phospholipid. Virions possess an internal lipid membrane that lies between the DNA core and the viral capsid, and, in some virions, an outer viral envelope. The origin of the internal lipid membrane is unclear. On one hand, the composition of the internal lipid membrane suggests that this membrane is not derived from host membranes but is produced de novo. However, by analogy to African swine fever virus, it has been suggested that the internal lipid membrane is derived from fragments of the endoplasmic reticulum and plays a key role in virion assembly. Viruses released from cells by budding acquire their outer envelope from the plasma membrane.
Carbohydrates
Carbohydrates are not present in purified virions.
Genome organization and replication
The replication strategy of iridoviruses is novel and has been elucidated primarily through the study of FV-3, the type species of the genus Ranavirus (Figure 5). Virion entry occurs by either receptor-mediated endocytosis (enveloped particles) or by uncoating at the plasma membrane (naked virions). Following uncoating, viral cores enter the nucleus where first stage DNA synthesis and the synthesis of immediate early (IE) and delayed early (DE) viral transcripts takes place. In a poorly understood process, one or more virion associated proteins act as transactivators and re-direct host RNA polymerase II to synthesize IE and DE viral mRNAs using the methylated viral genome as template. Gene products encoded by IE and DE viral transcripts include both regulatory and catalytic proteins. One of these gene products, the viral DNA polymerase, catalyzes the first stage of viral DNA synthesis. In this process, the parental viral genome serves as the template and progeny DNA that is genome-length, to at most twice genome length, is produced. Newly-synthesized viral DNA may serve as the template for additional rounds of DNA replication or early transcription, or it may be transported to the cytoplasm where the second stage of viral DNA synthesis occurs. In the cytoplasm, viral DNA is synthesized into large, branched concatamers that serve as the template for DNA packaging.
Viral DNA methylation also likely occurs in the cytoplasm and, although its precise role is uncertain, it is thought to protect viral DNA from endonucleolytic attack. Synthesis of late (L) viral transcripts occurs in the cytoplasm and full L gene transcription requires prior DNA synthesis. Homologs of the two largest subunits of RNA polymerase II are encoded by all iridoviruses. Whether these function only in the cytoplasm to transcribe L viral transcripts, or whether they also play a role in continued early transcription has not yet been determined. Virion formation takes place in the cytoplasm within morphologically distinct virus assembly sites. Within assembly sites concatameric viral DNA is packaged into virions, it is believed, via a “headful” mechanism that results in the generation of circularly permuted and terminally redundant genomes similar to those seen with the Enterobacteria phages T4 or P22. The degree of terminal redundancy varies from approximately 5 to 50%. Following assembly, virions accumulate in the cytoplasm within large paracrystalline arrays or acquire an envelope by budding from the plasma membrane. In the case of most vertebrate iridoviruses, the majority of virions remain cell-associated (Figure 5).
Antigenic properties
The genera are serologically distinct from one another. In the genus Iridovirus there exists one main group of serologically interrelated species and others which have little sero-relatedness. Several amphibian isolates (e.g., Rana esculenta iridovirus, REIR) and piscine isolates (e.g. EHNV) show serological cross-reactivity with FV-3 (genus Ranavirus). Although antibodies prepared against virions are often non-neutralizing in vitro, studies with FV3-infected Xenopus laevis indicate a marked antibody response following secondary infection and the generation of protective neutralizing antibodies. Moreover, consistent with a protective antibody response, inactivated virus vaccines protect against disease mediated by RSIV.
Biological properties
Iridoviruses have been isolated from only poikilothermic animals, usually associated with damp or aquatic environments, including marine habitats. Iridovirus species vary widely in their natural host range and in their virulence. Transmission mechanisms are poorly understood for the majority of these viruses. Invertebrate iridoviruses may be transmitted by cannibalism, endoparasitic wasps or parasitic nematodes. Viruses may be transmitted experimentally by injection or bath immersion, and naturally by co-habitation, feeding, or wounding. While many of these viruses cause serious, life-threatening infections, subclinical infections are common.
Genus Iridovirus
Type species Invertebrate iridescent virus 6
Distinguishing features
Particle diameter is 120–130 nm in ultrathin section. IIV-1 and IIV-2 are assumed to contain 1472 capsomers arranged in 20 trimers and 12 pentamers. A detailed description of the morphology of IIV-6 is presented in the introductory section on virion morphology.
Virions have an Mr of approximately 1.28×109, a buoyant density of 1.30–1.33 g cm−3, and a sedimentation coefficient S20,w of 2020–2250S. IIV-6 is sensitive to chloroform, SDS, sodium deoxycholate, ethanol, pH3 and pH11, but is not sensitive to trypsin, lipase, phospholipase A2 or EDTA. The sensitivity of IIV-6 to ether differs depending on whether an in vivo or in vitro assay is used to determine residual infectivity.
The unit length genome size of IIV-6 is 212,482 bp. The G+C content is typically 29–32%.Comparative genomic analysis of the IIV-6 and IIV-3 genomes shows no co-linearity between these two viral isolates.
Genetic studies have indicated the presence of discrete complexes of inter-related viruses within this genus: one large complex containing 10 viruses that may be candidates for new species, and two smaller complexes. Serological relationships follow a similar pattern.
Iridoviruses have been isolated from a wide range of arthropods, particularly insects in aquatic or damp habitats. Patently infected animals and purified viral pellets display violet, blue or turquoise iridescence. Non-apparent, non-lethal infections may be common in certain hosts. No evidence exists for transovarial transmission and where horizontal transmission has been demonstrated, it is usually by cannibalism or predation of infected invertebrate hosts. Following experimental injection, many members of the genus can replicate in a large number of insects. In nature, the host range appears to vary but there is evidence, for some viruses, of natural transmission across insect orders and even phyla. Invertebrate iridescent viruses have a global distribution.
Species demarcation criteria in the genus
The following species-defining characteristics and associated limits are preliminary in nature. The following definitions assume that all material being compared has been grown under near identical conditions and prepared for examination following identical protocols. It is recommended that members of both recognized virus species be included in all characterization studies of novel isolates.
- Amino acid sequence analysis of the MCP: PCR primers have been designed for conserved regions of this gene. Although the complete IIV-6 genome has been determined and the sequence of a number of other proteins from different isolates has been ascertained, this information has not been used for species differentiation and quantitative limits of similarity have not been established.
- DNA-DNA dot-blot hybridization: Hybridization values should be less than 50% for members of distinct species.
- RFLP: Using a panel of not fewer than four restriction endonucleases (both rare and frequent cutters) distinct species should show completely distinct restriction endonuclease profiles.
- Serology: Antisera from members of strains within a species should exhibit high levels of cross reactivity. Within and among species, comparison by Western blot analysis using antibodies raised against disrupted virions is the preferred method. Comparisons should be performed simultaneously wherever possible and reference species should be included in each determination.
The MCP of IIV-1 shows 66.4% aa sequence identity to that of IIV-6 and approximately 50% or lower aa sequence identity to iridoviruses in other genera. Less than 1% DNA–DNA hybridization for genomic DNA was detected by dot-blot method between IIV-1 and IIV-6 (stringency: 26% mismatch). Restriction endonuclease profiles (HindII, EcoRI, SalI) showed a coefficient of similarity of <66% between IIV-1 and IIV-6. These species did not share common antigens when tested by tube precipitation, infectivity neutralization, reversed single radial immunodiffusion or enzyme-linked immunosorbent assay. Genome and protein size differences are not useful in differentiating these species. The size of the MCP is well conserved among species.
List of species in the genus Iridovirus
| Invertebrate iridescent virus 1 | ||
| Invertebrate iridescent virus 1 | [M33542, M62953] | (IIV-1) |
| (Tipula iridescent virus) |
| (TIV) |
| Invertebrate iridescent virus 6 | ||
| Invertebrate iridescent virus 6 | [AF303741] | (IIV-6) |
| (Chilo iridescent virus) |
| (CIV) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Iridovirus but have not been approved as species
| Anticarsia gemmatalis iridescent virus | [AF042343] | (AGIV) |
| Invertebrate iridescent virus 2 | [AF042335] | (IIV-2) |
| (Sericesthis iridescent virus) |
|
|
| Invertebrate iridescent virus 9 | [AF025774] | (IIV-9) |
| (Wiseana iridescent virus) |
|
|
| Invertebrate iridescent virus 16 | [AF025775] | (IIV-16) |
| (Costelytra zealandica iridescent virus) |
|
|
| Invertebrate iridescent virus 21 |
| (IIV-21) |
| (Heliothis armigera iridescent virus) |
|
|
| Invertebrate iridescent virus 22 | [AF042341; M32799] | (IIV-22) |
| (Simulium sp. iridescent virus) |
|
|
| Invertebrate iridescent virus 23 | [AF042342] | (IIV-23) |
| (Heteronychus arator iridescent virus) |
|
|
| Invertebrate iridescent virus 24 | [AF042340] | (IIV-24) |
| (Apis iridescent virus) |
|
|
| Invertebrate iridescent virus 29 | [AF042339] | (IIV-29) |
| (Tenebrio molitor iridescent virus) |
|
|
| Invertebrate iridescent virus 30 | [AF042336] | (IIV-30) |
| (Helicoverpa zea iridescent virus) |
|
|
| Invertebrate iridescent virus 31 | [AF042337; AJ279821] | (IIV-31) |
| (Isopod iridescent virus) |
|
|
Genus Chloriridovirus
Type species Invertebrate iridescent virus 3
Distinguishing features
Particle diameter is approximately 180 nm in ultrathin section. The trimers and pentamers of invertebrate iridescent virus 3 (IIV-3) are larger than the corresponding structures of the genus Iridovirus, with probably 14 capsomers to each edge of the trimer. Particle size has historically been used to define viruses that are members of this genus, but the validity of that characteristic is uncertain.
Virions have a Mr of approximately 2.49–2.75 ×109, a buoyant density of approximately 1.354 g cm−3 in CsCl, and a S20,w of 4440–4460S. Infectivity is believed not to be sensitive to ether.
The genome size of IIV-3 is 190,132 bp with a G+C content of 48%. Of the 126 predicted genes, 27 have homologs in other sequenced iridovirids. IIV-3 and IIV-6 share 52 predicted genes that are not found in vertebrate iridovirids, including DNA topoisomerase II, NAD-dependent DNA ligase, SF1 helicase, IAP and a BRO protein. Thirty-three IIV-3 genes lack homologs in other iridovirids. No colinearity has been observed between IIV-3 and the genome of any other iridovirid sequenced to date.
The low levels of amino acid identity of predicted proteins to iridovirid homologs and phylogenetic analyses of conserved proteins indicate that IIV-3 is only distantly related to other iridovirid genera.
IIV-3 is serologically distinct from members of other genera.
IIV-3 is the only virus characterized from this genus, although more than 20 host species were reported with latent infections world-wide. Chloriridovirus-like infections have only been reported from Diptera with aquatic larval stages, mainly mosquitoes. There is evidence for transovarial transmission in mosquitoes infected by IIV-3. Horizontal transmission is achieved by cannibalism or predation of infected mosquitoes of other species. Patently infected larvae and purified pellets of virus iridesce, usually with a yellow-green color, although orange and red infections are known. IIV-3 appears to have a narrow host range compared to most members of the genus Iridovirus.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Chloriridovirus
| Invertebrate iridescent virus 3 | ||
| Invertebrate iridescent virus 3 | [AJ312708] | (IIV-3) |
| (Mosquito iridescent virus) |
|
|
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Chloriridovirus but have not been approved as species
None reported.
Genus Ranavirus
Type species Frog virus 3
Distinguishing features
Ranaviruses infect one or more members of the classes Reptilia, Amphibia and Osteichthyes. With the exception of SGIV and GIV they encode a cytosine DNA methyltransferase.
Particle diameter is approximately 150 nm in ultrathin section. Enveloped virions, released by budding, measure 160–200 nm in diameter. The capsid has a skewed symmetry with T=133 or 147. The internal lipid bilayer likely contains transmembrane proteins. The nucleoprotein core consists of a long coiled filament 10 nm wide.
Buoyant density is 1.28 g cm−3 for enveloped particles and 1.32 g cm−3 for nonenveloped particles. Infectivity is rapidly lost at pH 2.0–3.0 and at temperatures above 50 °C. Particles are inactivated by treatment with ether, chloroform, sodium deoxychlorate, and phospholipase A.
Virions contain a single linear dsDNA molecule. The genome is circularly permuted and approximately 30% terminally redundant. The unit genome size is approximately 105 kbp with a G+C content of about 54% (Table 1). Cytosines within the dinucleotide sequence CpG are methylated by a virus-encoded cytosine DNA methyltransferase. DNA methylation likely occurs in the cytoplasm and may be important in protecting DNA from viral endonucleases.
The replication cycle of FV-3 serves as the model for the family and has been discussed above (Figure 5). The complete genomes of seven ranaviruses (SGIV, GIV, EHNV, STIV, TFV, FV-3 and ATV) have been sequenced and, while possessing homologous proteins, they are not, for the most part, co-linear (Table 1, Figure 4). Based on whole genome dot plot comparisons there are three genomic phenotypes among the seven completely sequenced ranaviruses (FV3/TFV/STIV, ATV/EHNV, SGIV/GIV). Dot plot analyses shows that the FV3- and ATV-like viruses are much more closely related to each other than are either of these two types to SGIV-like viruses.
Ranaviruses such as FV-3 are serologically and genetically distinct from members of other genera. However, several piscine, reptilian and amphibian ranavirus isolates show serological cross-reactivity with FV-3. Serological cross-reactivity likely reflects the marked amino acid sequence conservation (i.e., >90% identity) within the MCP and other viral proteins.
Viral transmission occurs by feeding, parenteral injection, or environmental exposure. Ranaviruses grow in a wide variety of cultured fish, amphibian and mammalian cells at temperatures up to 32 °C. Infection causes marked cytopathic effects culminating in cell death, likely by apoptosis, such as the mitochondrion-mediated apoptosis observed in fish cells infected with Rana grylio virus (RGV). In contrast to their marked pathogenicity in vitro, their effect in animals depends on the viral species, and on the identity and age of the host animal. For example, largemouth bass virus (LMBV) shows evidence of widespread infection in the wild, but is only rarely linked to serious disease. Likewise, FV-3 infection leads to death in tadpoles, but often causes only non-apparent infections in adults that resolve within two weeks. In contrast, RGV causes a lethal syndrome in Rana gyrlio tadpoles and young adults. It is likely that environmental stress leading to immune suppression increases the pathogenicity of a given ranavirus. Ranavirus infections are often not limited to a single species or taxonomic class of animals. For example, EHNV has been reported to infect at least 13 species of fish. In addition, BIV, a highly virulent pathogen of the burrowing frog Lymnodynastes ornatus, can be experimentally transmitted to fish. Therefore, isolation of a ranavirus from a new host species does not necessarily identify a new viral species. In their most severe clinical manifestations, ranaviruses such as FV-3, ATV, European catfish virus (ECV) and EHNV are associated with systemic disease and show marked hemorrhagic involvement of internal organs such as the liver, spleen, kidneys and gut.
Species demarcation criteria in the genus
Ranaviruses cause systemic disease in fish, amphibians and reptiles. Members of the six viral species are differentiated from one another by multiple criteria: RFLP profiles, virus protein profiles, DNA sequence analysis and host specificity. PCR primers have been designed to amplify 3’ and 5’ regions within the MCP gene for identification purposes. Definitive quantitative criteria based on the above features have not yet been established to delineate different viral species. Generally, if a given isolate shows a distinct RFLP profile, possesses a distinctive host range and is markedly different from other viruses at the aa sequence level, it is considered a distinct viral species. For example, ranavirus DNA digested with KpnI can be ordered into several groups based on RFLP profiles. Strains within the same species shared >70% of their bands in common and showed >95% aa sequence identity within the MCP or other key genes (e.g., ATPase, eIF-2α homolog).
List of species in the genus Ranavirus
| Ambystoma tigrinum virus |
|
|
| Ambystoma tigrinum virus | [AY150217] | (ATV) |
| (Regina ranavirus) |
|
|
| Bohle iridovirus |
|
|
| Bohle iridovirus | [AF157650, AF157651] | (BIV) |
| Epizootic hematopoietic necrosis virus |
|
|
| Epizootic hematopoietic necrosis virus | [FJ433873] | (EHNV) |
| European catfish virus |
|
|
| European catfish virus | [AF157678, AF127911] | (ECV) |
| (European sheatfish virus) |
| (ESV) |
| Frog virus 3 |
|
|
| Frog virus 3 | [AY548484] | (FV-3) |
| (Tiger frog virus) | [AF389451] | (TFV) |
| Santee-Cooper ranavirus |
|
|
| Santee-Cooper ranavirus | [AF080250] | (SCRV) |
| (Largemouth bass virus) |
| (LMBV) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Ramavirus but have not been approved as species
| Rana esculenta iridovirus |
| (REIR) |
| Singapore grouper iridovirus | [AY521625] | (SGIV) |
| Grouper iridovirus |
| (GIV) |
| Testudo iridovirus |
| (ThIV) |
| Rana catesbeiana virus-Z |
| (RCV-Z) |
Genus Lymphocystivirus
Type species Lymphocystis disease virus 1
Distinguishing features
Particle size varies from 198 to 227 nm for lymphocystis disease virus 1 (LCDV-1) and 200 nm for LCDV-2. The capsid may show a fringe of fibril-like external protrusions about 2.5 nm in length and a double-layered outer envelope.
Virions are heat labile. Infectivity is sensitive to treatment with ether or glycerol.
The genome length of LCDV-1 is 102.6 kbp and that of LCDV-C is 186 kbp. LCDV-C possesses the largest genome among known members within the three genera of vertebrate iridoviruses. Contour length measurements for LCDV-1 were determined by electron microscopy and indicate the DNA molecule to be 146 kbp; the degree of terminal redundancy is approximately 50% but varies considerably among virions. The G+C content is 29.1% for LCDV-1 and 27.2% for LCDV-C. Like FV-3, the genome is highly methylated. The presence of 5-methylcytosine occurs at 74% of CpG, 1% of CpC and 2–5% of CpA giving an overall level of methylation of 22%. The complete DNA sequence is known for LCDV-1 and LCDV-C.
The LCDV-1 genome contains 108 largely non-overlapping ORFs, 38 of which show significant homology to proteins of known function. These 38 ORFs represent 43% of the coding capacity of the genome. The presence of a DNA methyltransferase and a methyl-sensitive restriction endonuclease with specificity for a CCGG target site may be indicative of a restriction-modification system capable of degrading host genomic DNA while protecting viral DNA by specific methylation. LCDV-1 DNA contains numerous short direct, inverted and palindromic repetitive sequence elements. The LCDV-C genome contains 178 non-overlapping ORFs, 103 of which are homologs to the corresponding ORFs of LCDV-1 and 75 potential genes that were not found in LCDV-1 or in other iridovirids. Among these 75 genes, there are eight genes that contain conserved domains of cellular genes and 67 novel genes that do not show any signicant homology with the sequences in public databases. Although LCDV-1 and LCDV-C possess some genes in common, their genomic organization (i.e. gene order) is markedly different. Furthermore, a large number of tandem and overlapping repeated sequences were observed in the LCDV-C genome. As expected, LCDV-C MCP is most similar to the MCP of LCDV-1. Lack of a suitable cell line has hindered studies of LCDV replication. Virus assembly occurs in and around virogenic stroma within viral assembly sites. Crescent-shaped capsid precursors develop into fully-formed capsids followed by condensation of the core structures.
LCDV-1 and LCDV-C infect flounder and plaice, whereas LCDV-2 infects dab. Infection results in benign, wart-like lesions comprising grossly hypertrophied cells occurring mostly in the skin and fins. The disease has been observed in over 100 teleost species although virus species other than LCDV may cause a similar clinical disease. The duration of infected cell growth and viral proliferation is highly variable (5 days to 9 months) and is likely temperature-dependent. Virions are released following degeneration of the lesions. Transmission is achieved by contact; external sites, including the gills, are the principal portals of entry. High host population densities and external trauma are believed to enhance transmission. Implantation and injection are also effective routes of transmission. The incidence of disease may be higher in the presence of certain fish ectoparasites. LCDV is generally not considered to be of major economic importance. However, although infections are usually benign and self-limiting, there may be commercial concerns because the warty appearance of infected fish leads to market rejection. Mortalities may occur, especially when infections involve the gills or when there is debilitation or secondary bacterial infection. LCDV-1, LCDV-C and LCDV-2 are difficult to culture in vitro although limited growth has been reported in several fish cell lines.
Species demarcation criteria in the genus
Definitive criteria have not yet been established to delineate the viral species. The species are distinguished from one another by host specificity, histopathology and molecular criteria: viral protein profiles, DNA sequence analysis and PCR. PCR primers targeted to regions within the MCP and ORF167L can be used to distinguish between species.
List of species in the genus Lymphocystivirus
| Lymphocystis disease virus 1 | ||
| Lymphocystis disease virus 1 | [L63545] | (LCDV-1) |
| (Flounder lymphocystis disease virus) |
| (FLDV) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Lymphocystivirus
| Lymphocystis disease virus 2 |
| (LCDV-2) |
| Lymphocystis disease virus-China | [AY380826] | (LCDV-C) |
| Lymphocystis disease rockfish virus | [AB213004, AB213005, AB213006] | (LCDV-RF) |
| Dab lymphocystis disease virus |
|
|
Genus Megalocytivirus
Type species Infectious spleen and kidney necrosis virus
Distinguishing features
Virons are similar in appearance to those of ranaviruses and genomes are highly methylated. Infections in vivo lead to the appearance of enlarged cells (inclusion body bearing cells) that are characteristic of megalocytivirus infections.
Virions possess icosahedral symmetry and are about 140–200 nm in diameter.
Virions are sensitive to heat (56 °C for 30 min), sodium hypochlorite, UV irradiation, chloroform and ether, and are variably inactivated by exposure to pH3 and pH11.
The complete genomes of ISKNV, RBIV, RSIV and orange spotted grouper iridovirus (OSGIV) have been sequenced. ISKNV virions contain a single, linear dsDNA molecule of 111,362 bp with a G+C content of 54.8%. As with other members of the family, genomic DNA is circularly permuted, terminally redundant and highly methylated.
Genomic sequence comparisons show co-linearity among all of the completely sequenced members of the genus. However, they do not share sequence co-linearity with other members of the family.
No serotypes are reported and all megalocytiviruses analyzed to date appear to be members of the same viral species or several closely related species. Polyclonal anti-RSIV serum shows cross-reactivity with ESV- and EHNV-infected cells, whereas monoclonal anti-RSIV antibodies react only with RSIV-infected cells. Megalocytiviruses show high levels (i.e. >93%) of aa sequence identity among the proteins characterized to date.
Iridoviruses infecting red seabream, mandarin fish and over 20 other species of marine and tropical fish have been known since the late 1980s. Isolates from red seabream (RSIV) and mandarin fish (ISKNV) have been studied extensively. Viral infection is characterized by the formation of inclusion body-bearing cells (IBC). IBCs may be derived from virus-infected macrophages and enlarge by the growth of a unique inclusion body that may be sharply delineated from the host cell cytoplasm by a limiting membrane. When a limiting membrane is seen, the inclusions contain the viral assembly site and possess abundant ribosomes, rough ER and mitochondria. IBCs frequently appear in the spleen, hematopoietic tissue, gills and digestive tract. Necrotized splenocytes are also observed. Transmission has been demonstrated by feeding, parenteral injection and by environmental exposure. Megalocytiviruses naturally infect and cause significant mortality in freshwater and marine fish in aquaculture facilities in China, Japan and SE Asia. A partial list of susceptible fish species includes mandarin fish (Siniperca chuatsi), red seabream (Pagrus major), grouper (Epinephelus spp.), yellowtail (Seriola quinqueradiata), striped beakperch (Oplegnathus fasciatus), red drum (Sciaenops ocellata) and African lampeye (Aplocheilichthys normani). The virus grows in several cultured piscine cell lines and causes a characteristic enlargement of infected cells. Outbreaks of disease caused by ISKNV occur only in fish cultured at temperatures >20 °C. A vaccine targeted to RSIV has been developed suggesting that infection/immunization is capable of eliciting protective antibodies.
Species demarcation criteria in the genus
Megalocytiviruses are distinguished from ranaviruses and lymphocystiviruses by their cytopathological presentation (i.e., inclusion body-bearing cells) and sequence analysis of key viral genes, e.g., ATPase and MCP, for which PCR primers have been developed. Megalocytiviruses show >94% sequence identity within these genes, whereas sequence identity with ranaviruses and lymphocystiviruses is <50%. Based on sequence analysis and serological studies, all megalocytiviruses isolated to date appear to be strains of the same or several closely-related viral species.
List of species in the genus Megalocytivirus
| Infectious spleen and kidney necrosis virus | ||
| Infectious spleen and kidney necrosis virus | [AF371960= NC_003494] | (ISKNV) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Megalocytivirus but have not been approved as species
| Red seabream iridovirus | [BD143114] | (RSIV) |
| Sea bass iridovirus | [AB043977] | (SBIV) |
| African lampeye iridovirus | [AB043979] | (ALIV) |
| Grouper sleepy disease iridovirus | [AB043978] | (GSDIV) |
| Dwarf gourami iridovirus |
| (DGIV) |
| Taiwan grouper iridovirus |
| (TGIV) |
| Rock bream iridovirus | [AY532606] | (RBIV) |
| Orange-spotted grouper iridovirus | [AY894343] | (OSGIV) |
| Turbot iridovirus |
| (TBIV) |
| Spotted knifejaw iridovirus | [GQ202216, GQ202217] | (SKIV) |
List of other related viruses which may be members of the family Iridoviridae but have not been approved as species
| White sturgeon iridovirus |
| (WSIV) |
| Erythrocytic necrosis virus |
| (ENV) |
Phylogenetic relationships within the family
Phylogenetic analysis using the 26 core iridovirus genes from the 14 completely sequenced iridoviruses shows a clear division between the genera within the family Iridoviridae (Figure 6). Members of genera Ranavirus, Lymphocystivirus and Megalocytivirus are in three separate clades with a common ancestor, while members of the genera Iridovirus and Chloriridovirus form two clades that are more distantly related. In addition, the phylogeny shows a distinct lineage for each genus within the family Iridoviridae, with the vertebrate genera being more closely related to each other as compared to the invertebrate genera.
Similarity with other taxa
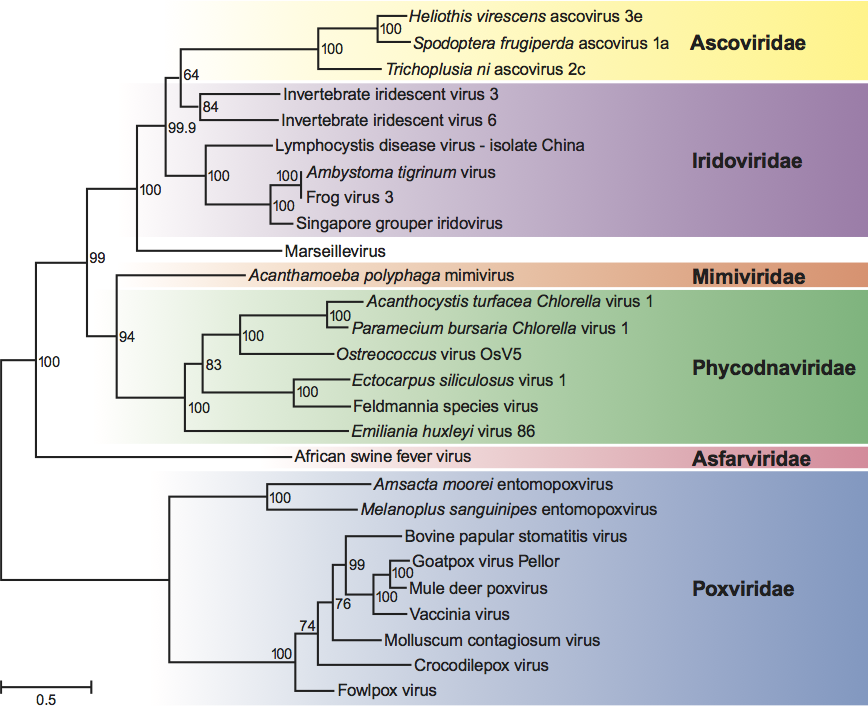
The iridovirus D5 ATPase, A32 ATPase, A1L/VLTF2 transcription factor, MCP and viral DNA polymerase B genes share aa sequence similarities to other nucleocytoplasmic large DNA viruses (NCLDV) such as African swine fever virus (Asfarviridae), species within the family Ascoviridae, Paramecium bursaria Chlorella virus 1 (Phycodnaviridae), Marseillevirus (Mimivirus), and poxviruses (Poxviridae) (Figure 7). Although not shown here, more distant relatedness to other NCLDV species including herpesviruses, adenoviruses and baculoviruses has also been noted.
Derivation of names
Chloro: from Greek chloros, “green”.
Cysti: from Greek kystis, “bladder/sac”.
Irido: from Greek iris, iridos, goddess whose sign was the rainbow, hence iridescent, “shining like a rainbow”, from the appearance of patently infected invertebrates and centrifuged pellets of virions.
Lympho: from Latin lympha, “water”.
Megalocyti: from the Greek, meaning “enlarged cell”.
Rana: from Latin rana, “frog”.
Further reading
Chinchar, V.G. (2002). Ranaviruses (family Iridoviridae): Emerging cold-blooded killers. Arch. Virol., 147, 447-470.
Chinchar, V.G., Hyatt, A., Miyazaki, T. and Williams, T. (2009) Family Iridoviridae: Poor viral relations no longer. Curr. Top. Microbiol. Immunol., 328, 123-170.
He, J.G., Deng, M., Weng, S.P., Li, Z., Zhou, S.Y., Long, Q.X., Wang, X.Z. and Chan, S.M. (2001). Complete genome analysis of the mandarin fish infectious spleen and kidney necrosis iridovirus. Virology, 291, 126-139.
Hyatt, A.D., Gould, A.R., Zupanovic, Z., Cunningham, A.A., Hengstberger, S., Whittington, R.J., Kattenbelt, J. and Coupar, B.E.H. (2000). Comparative studies of piscine and amphibian iridoviruses. Arch. Virol., 145, 301-331.
Jakob, N.J., Muller, K., Bahr, U. and Darai, G. (2001). Analysis of the first complete DNA sequence of an invertebrate iridovirus: Coding strategy of the genome of Chilo iridescent virus. Virology, 286, 182-196.
Jancovich, J.K., Bremont, M., Touchman, J.W. and Jacobs, B.L. (2010). Evidence for multiple recent host shifts among the ranaviruses (family Iridoviridae). J. Virol., 84, 2636-2647.
Morales, H.D. and Robert, J. (2007). Characterization of primary and memory CD8 T-cell responses against ranavirus (FV3) in Xenopus laevis. J. Virol., 81, 2240-2248.
Nakajima, K. and Kurita, J. (2005). Red sea bream iridovirus. Virus, 55, 115-126.
Sudthongkong, C., Miyata, M. and Miyazaki, T. (2002). Viral DNA sequences of genes encoding the ATPase and the major capsid protein of tropical iridovirus isolates which are pathogenic to fishes in Japan, South China Sea, and Southeast Asian countries. Arch. Virol., 47, 2089-2109.
Williams, T., Barbosa-Solomieu, V. and Chinchar, V.G. (2005). A decade of advances in iridovirus research. Adv. Virus Res., 65, 173-248.
Contributed by
Jancovich, J.K., Chinchar, V.G., Hyatt, A., Miyazaki, T., Williams, T. and Zhang, Q.Y.
Figures
Figure 1 (Top left) Outer shell of invertebrate iridescent virus 2 (IIV-2) (from Wrigley et al. (1969). J. Gen. Virol., 5, 123; with permission). (Top right) Schematic diagram of a cross-section of an iridovirus particle, showing capsomers, transmembrane proteins within the lipid bilayer, and an internal filamentous nucleoprotein core (from Darcy-Tripier et al. (1984). Virology, 138, 287; with permission). (Bottom left) Transmission electron micrograph of a fat head minnow cell infected with an isolate of European catfish virus. Nucleus (nu); virus inclusion body (VIB); paracrystalline array of non-enveloped virus particles (arrows); incomplete nucleocapsids (arrowheads); cytoplasm (cy); mitochondrion (mi); the bar represents 1 m (from Hyatt et al. (2000). Arch. Virol., 145, 301; with permission). (insert) Transmission electron micrograph of particles of frog virus 3 (FV-3), budding from the plasma membrane. Arrows and arrowheads identify the viral envelope; the bar represents 200 nm
(from Devauchelle et al. (1985). Curr. Topics Microbiol. Immunol., 116, 1; with permission.)
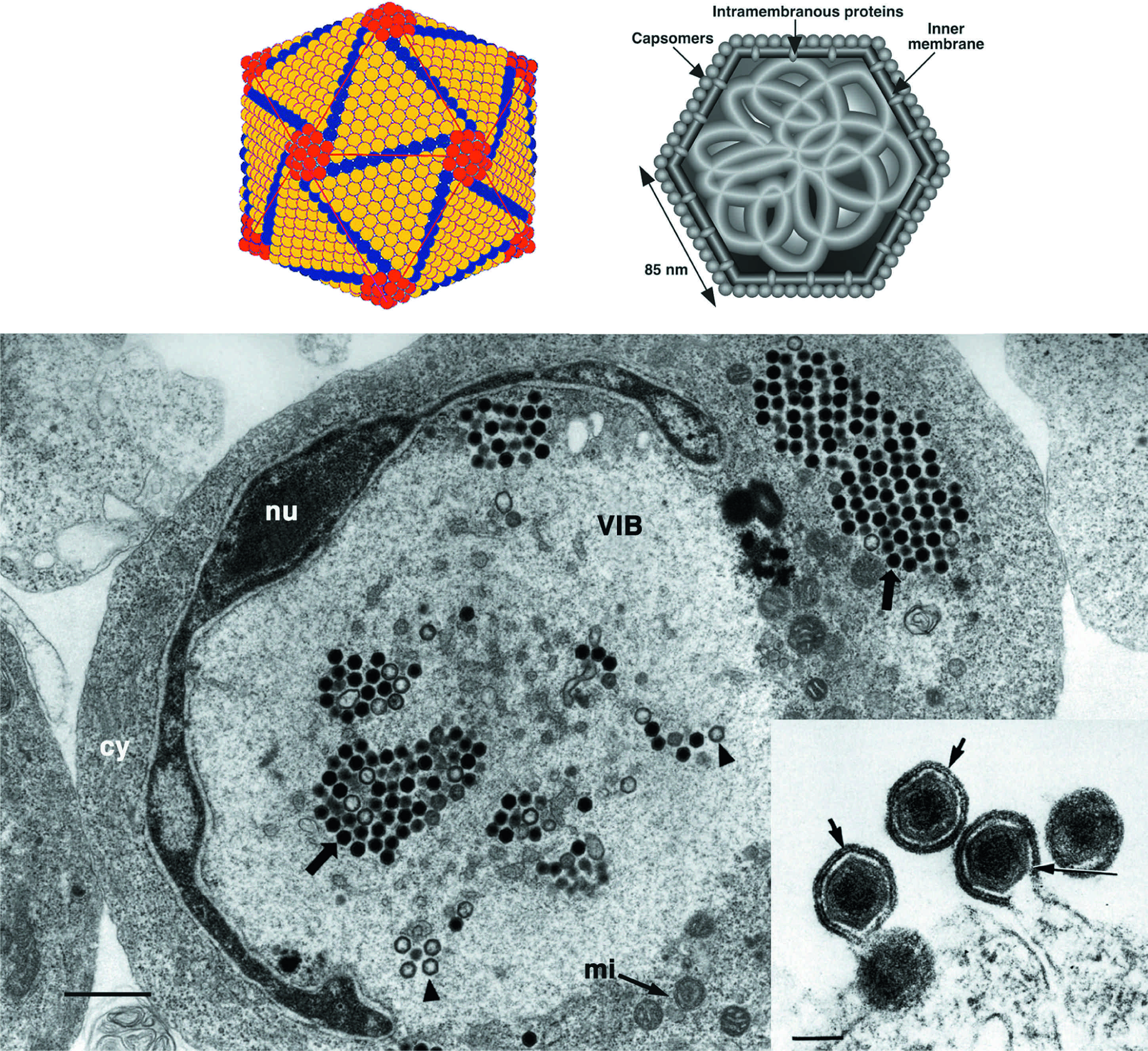
Figure 2 Three-dimensional reconstruction of an IIV-6 particle at a resolution of 1.3 nm. (A) 3-D density map of particle viewed along an icosahedral two-fold axis. The external fringe of filaments has been blurred away due to their varying position in relation to the symmetry of the capsid. (B) Central cross-section, one pixel thick. A lipid bilayer follows the inner contour and icosahedral symmetry of the capsid shell whereas the core appears to lack any structures that are arranged following icosahedral symmetry. (C) Magnified view of the central region outlined in (A). The diagonal white line indicates the cleavage plane between adjacent trisymmetrons. (D) Magnified view of the propeller-like pentamer complex at the five-fold vertex. This complex differs from the trimeric capsomers in that it is larger, has a small axial hole, and lacks an external fibre. (E) Magnified view of boxed region in (B) showing transverse section of the pentamer complex and adjacent trisymmetrons. (F) Same as (E) but with the pentamer complex, three capsomers, and their fibres individually outlined. (G) Transmembrane anchor proteins beneath the pentasymmetrons showing two sticklike entities. The longer of the two (long arrow) crosses both leaflets of the bilayer, whereas the other (short arrow) stops at the outer leaflet of the internal lipid membrane.
(From Yan et al. (2009). J. Mol. Biol., 385, 12871299; with permission.)
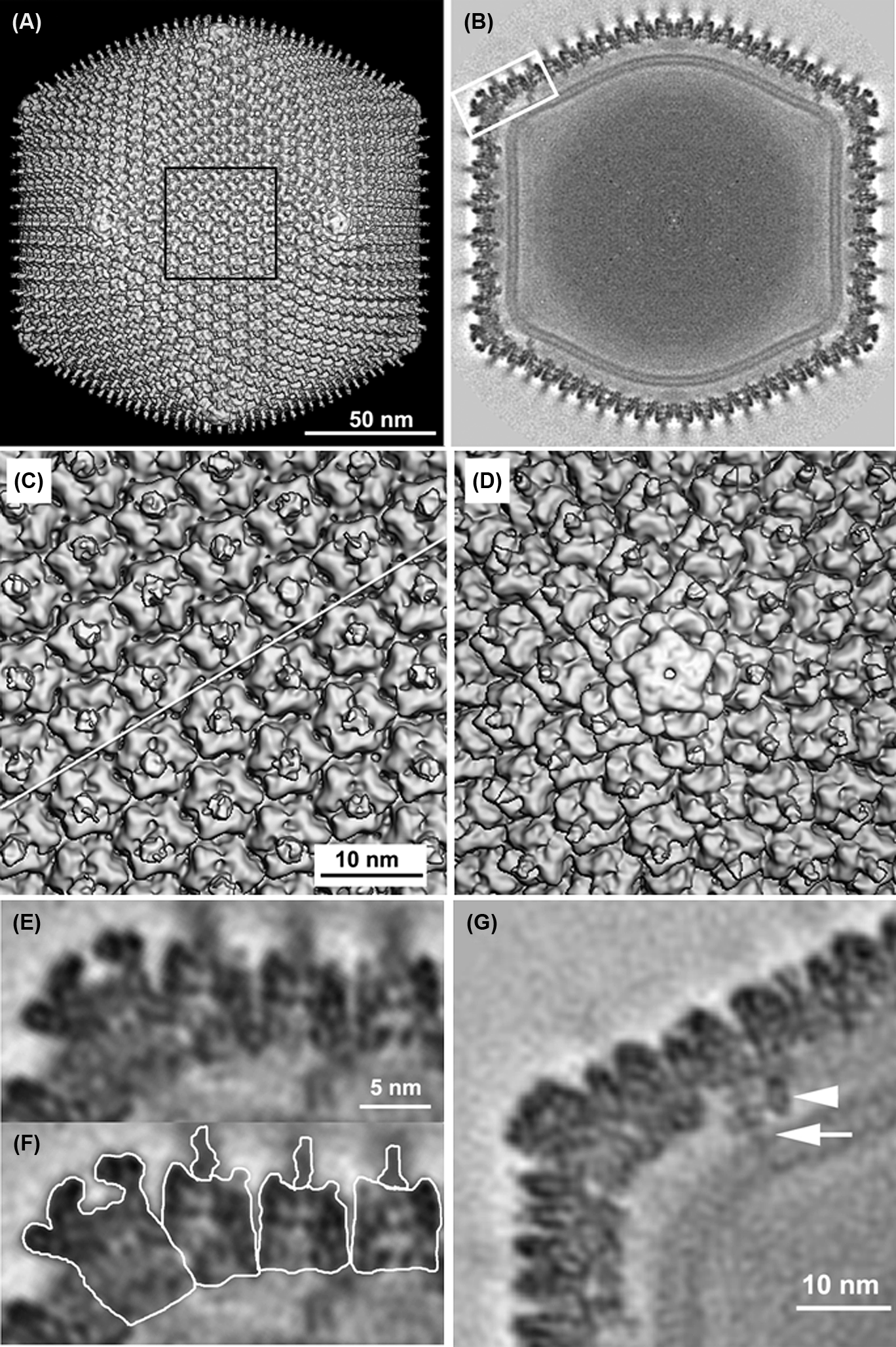
Figure 3 Schematic planar diagram of the IIV-6 capsid structure with the central pentamer complex (white pentagon) omitted for clarity. The centrally located white ellipse, triangle and pentagon symbols highlight the positions of 2-, 3-, and 5-fold icosahedral axes, respectively. All major capsid protein trimers are shown as three small grey disks enclosed by a hexagon. The icosahedral asymmetric unit contains 24 and one-third of these capsomers (numbered 125). Each trisymmetron contains 55 capsomers, all oriented similarly and rotated by 60 relative to those in the adjacent trisymmetron. Finger proteins bind to each trisymmetron (nine within one asymmetric unit are numbered in white), a total of 27 in each trisymmetron. A total of 18 Zip dimers are present at the interface between one trisymmetron and its adjacent trisymmetrons. Zip monomers are located at the interface between a trisymmetron and its neighbouring pentasymmetrons. The transmembrane anchor proteins (shown in Figure 2G) are located under capsomers 2 and 3 beneath the pentasymmetron.
(From Yan et al. (2009). J. Mol. Biol., 385, 12871299; with permission.)
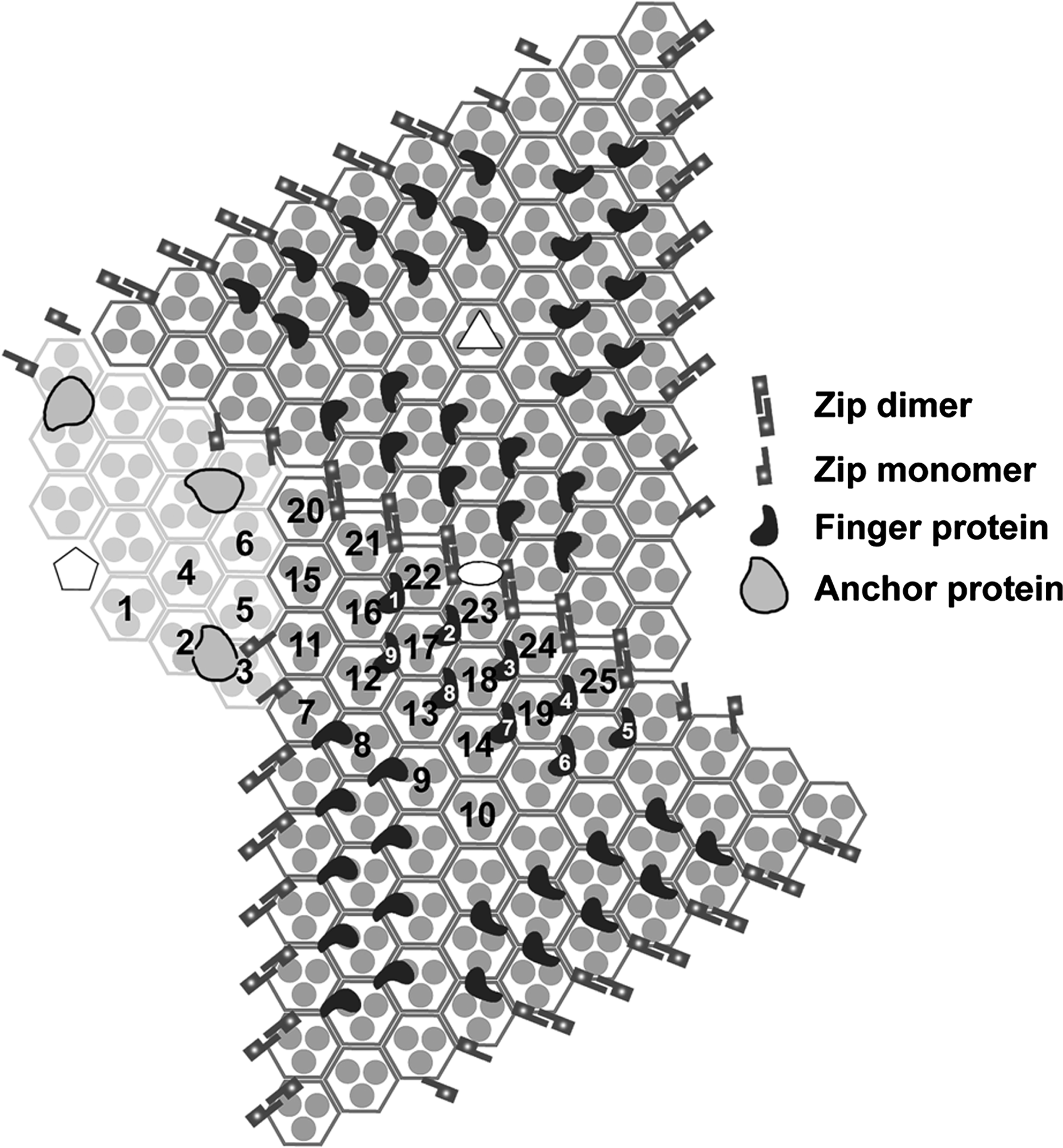
Figure 4 Genomic structure of Ambystoma tigrinum virus (ATV). Arrows represent viral ORFs with their size, position and orientation shown. ORFs of known function are colored in red and their putative proteins identified; ORFs with known homology to tiger frog virus (TFV) are in blue; and those of unknown function or with no homology to TFV are indicated in black.
(From Jancovich et al. (2003). Virology, 316, 90103; with permission.)
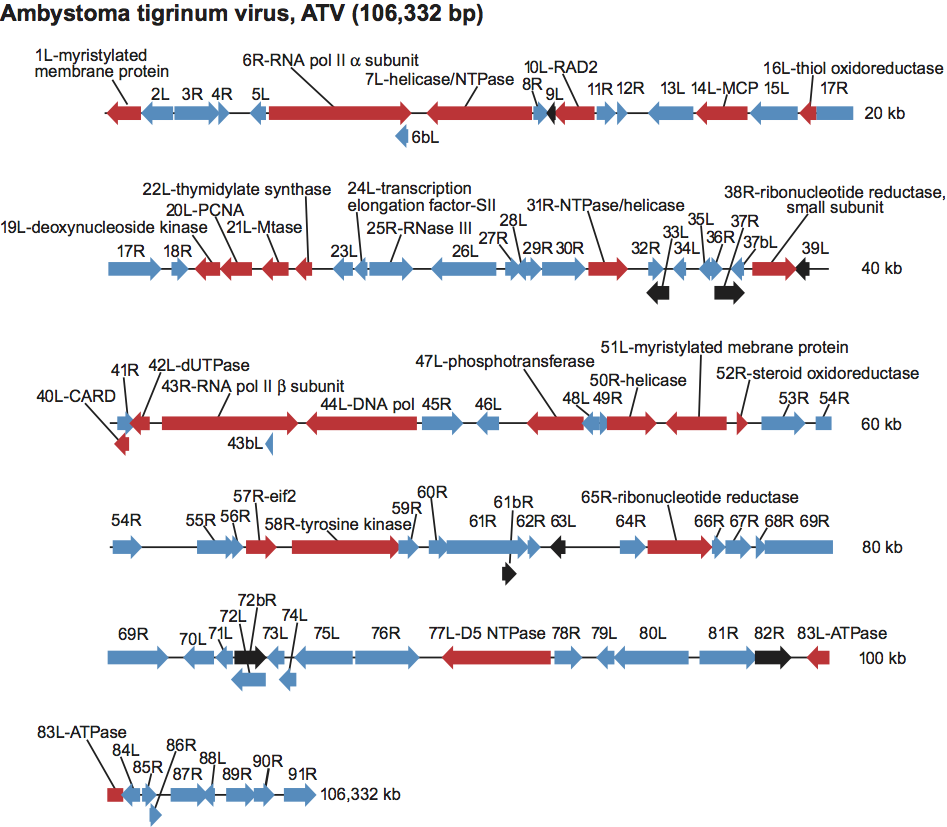
Figure 5 Replication cycle of frog virus 3 (FV-3).
(From Chinchar et al., (2002). Arch. Virol., 147, 447; with permission.)
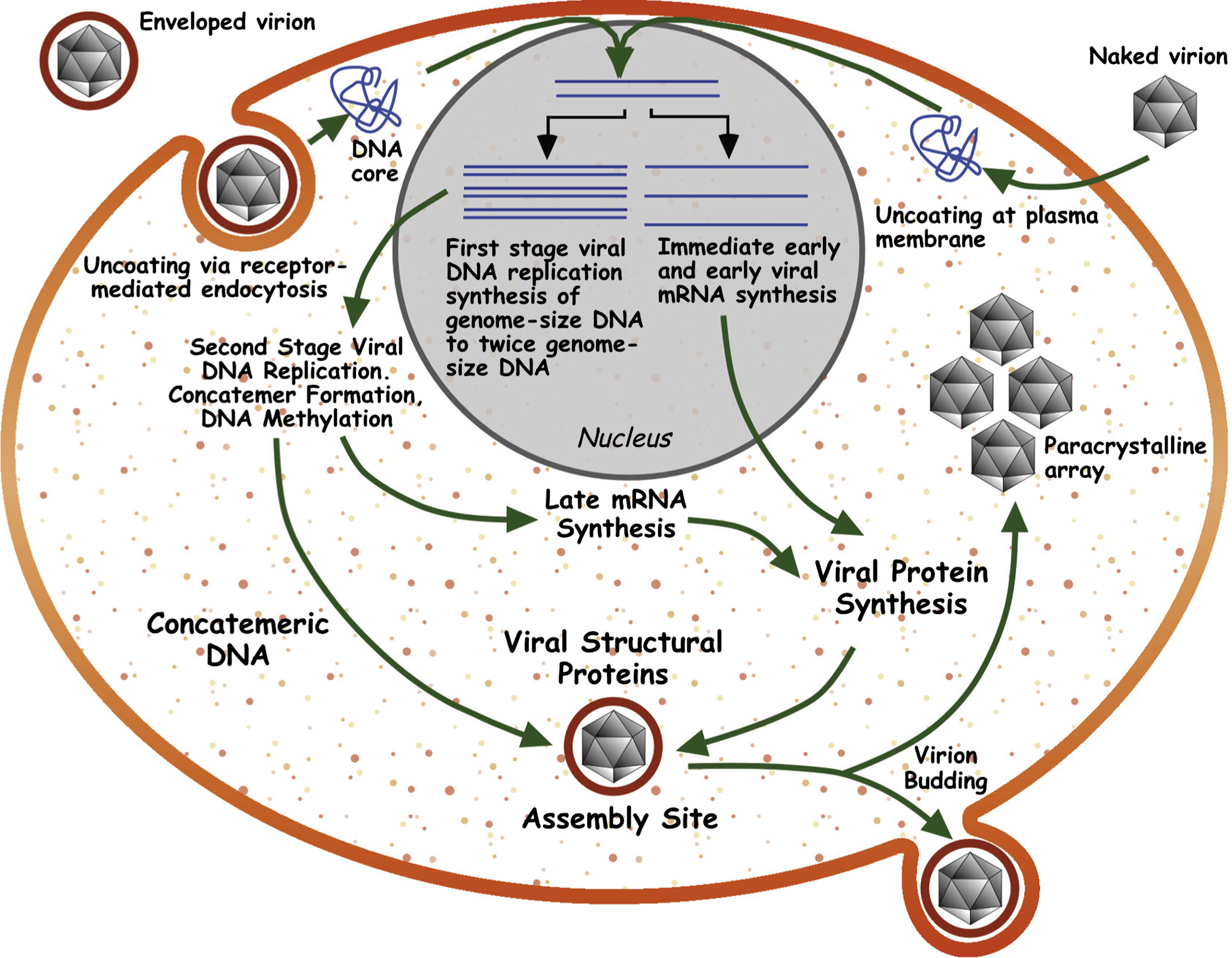
Figure 6 Concatenated phylogeny of 26 conserved iridovirid sequences. Phylogenetic relationship of 26 conserved ORFs from the 14 completely sequenced iridovirid genomes. The neighbor-joining tree obtained using MEGA4 is shown with the statistical support indicating the robustness of the inferred branching pattern as assessed using the bootstrap test.
(Modified from Jancovich et al. (2010). J. Virol., 84, 26362647; with permission.)
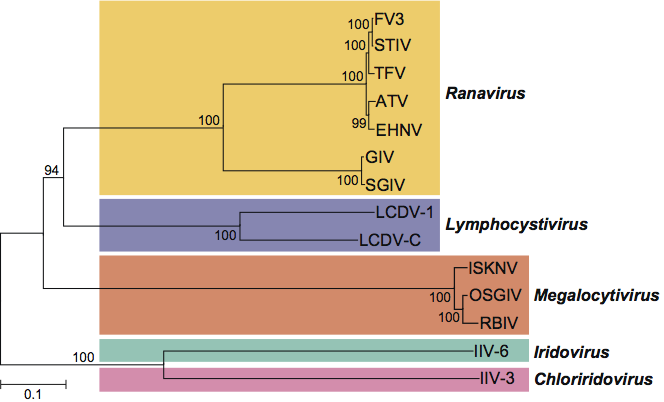
Figure 7 A maximum-likelihood tree based on concatenated alignments (1849 positions) of five nucleocytoplasmic large DNA viruses (NCLDV) core proteins: D5 type ATPase, DNA polymerase B, A32 ATPase, major capsid protein and A1L/VLTF2 transcription factor. The tree was built using TreeFinder.
(From Boyer et al. (2009). PNAS, 106, 2184821853; with permission.)