Order: Herpesvirales
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions have complex and characteristic structures consisting of both symmetrical and non-symmetrical components. The spherical virion is comprised of the core, capsid, tegument and envelope (Figure 1). The core consists of the viral genome packaged as a single, linear, dsDNA molecule into a preformed capsid. DNA is packed in a liquid-crystalline array that fills the entire internal volume of the capsid. The mature capsid is a T=16 icosahedron. In virions of human herpesvirus 1 (HHV-1), the 16 nm thick protein shell has an external diameter of 125 nm. The 11 pentons and 150 hexons (a total of 161 capsomers) are composed primarily of five and six copies, respectively, of the same protein and are joined by masses, termed triplexes, which are made of two smaller proteins present in a 2:1 ratio (Figure 1). The twelfth pentonal position is occupied by a ring-like structure consisting of 12 copies of the capsid portal protein.
Capsids assemble by co-condensation around a protein scaffold to form a procapsid in which the subunits are weakly connected. Proteolytic cleavage of the scaffolding protein triggers loss of scaffold and reorganization of the shell into the characteristic capsid form. The structure of the tegument is poorly defined, with evidence of symmetry only in the region immediately adjacent to the capsid. Electron tomography indicates that there are inner (capsid-associated) and outer (envelope-associated) tegument layers, and that capsids may be non-symmetrically situated within the envelope. The tegument contains many proteins, not all of which are required for the formation of virions. Individual tegument proteins can vary markedly in abundance. Enveloped tegument structures lacking capsids can assemble and are released from cells along with virions. The envelope is a lipid bilayer that is intimately associated with the outer surface of the tegument. It contains a number (at least 11 in HHV-1) of different integral viral glycoproteins that form a network of closely spaced spikes (mean of 659 per HHV-1 virion) of at least three distinct morphologies.
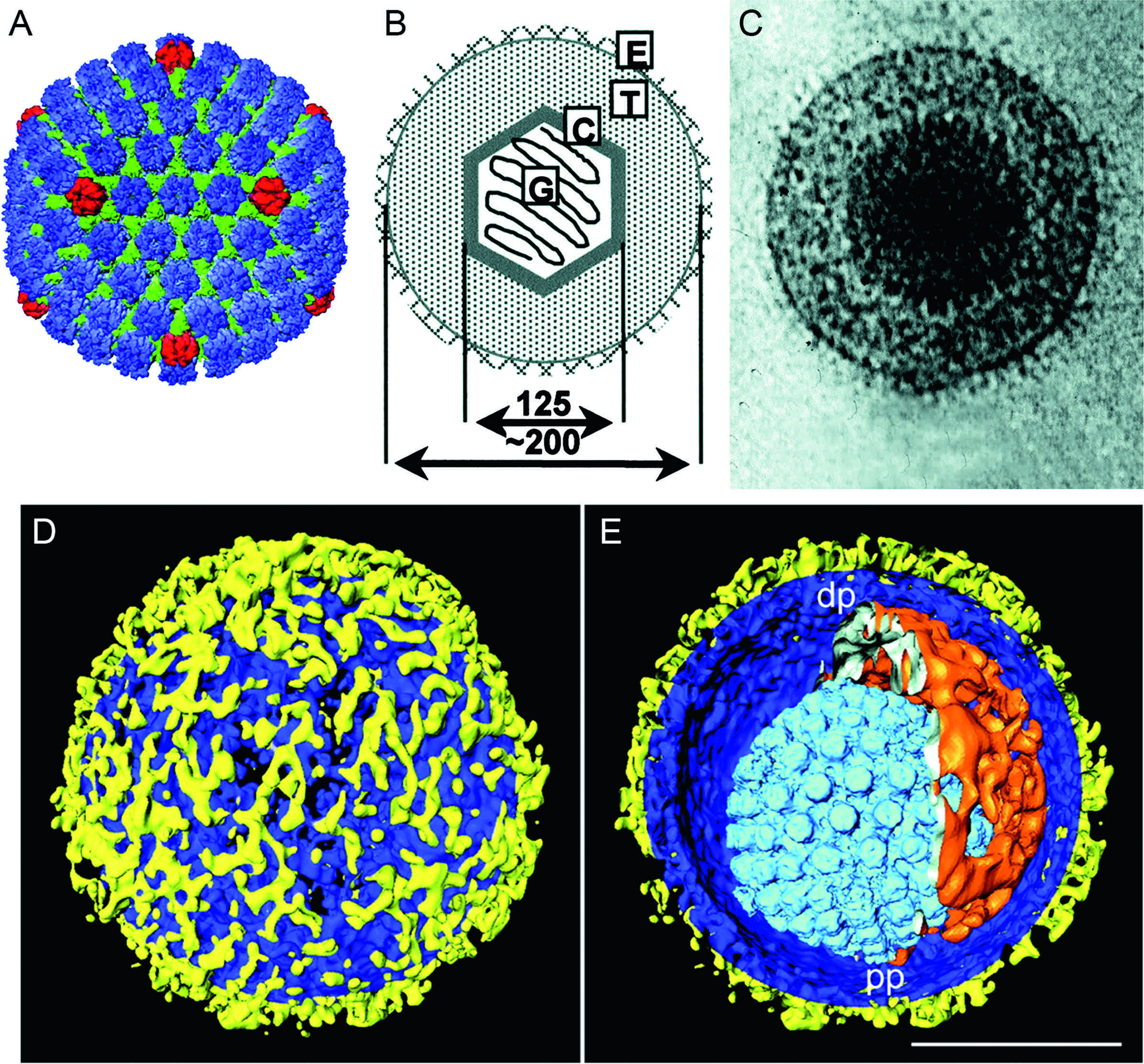
Figure 1. Herpesvirus morphology. (A) Reconstruction of a human herpesvirus 1 (HHV-1) capsid generated from cryo-electron microscope images, viewed along the two-fold axis. The hexons are shown in blue, the pentons in red and the triplexes in green (courtesy of W. Chiu and H. Zhou; Zhou, Z.H., Dougherty, M., Jakana, J., He, J., Rixon, F.J. and Chiu, W. (2000). Seeing the herpesvirus capsid at 8.5 Å. Science, 288, 877–880; reprinted with permission from AAAS).
Physicochemical and physical properties
The mass of the HHV-1 virion is about 13×10−16 g, of which the DNA comprises about 10%. The mass of a full capsid is about 5 ×10−16 g. The buoyant density of virions in CsCl is 1.22–1.28 g cm−3. The stability of different herpesviruses varies considerably, but they are generally unstable to desiccation and low pH. Infectivity is destroyed by lipid solvents and detergents.
Nucleic acid
The genomes are composed of linear dsDNA ranging from 125 to 295 kbp in size and from 32 to 75% in G+C content. The genomes examined in sufficient detail contain a single nucleotide extension at the 3′-ends, and no terminal protein has been identified. The arrangement of reiterated sequences (direct or inverted, at the termini or internally) results in a number of different genome structures (Figure 2), including isomers that result from recombination between inverted terminal and internal reiterations.
In Figure 2, structure 1 shows a unique sequence flanked by a direct repeat that may be larger than 10 kbp (human herpesvirus 6; HHV-6) or as short as 30 bp (murid herpesvirus 1; MuHV-1). Structure 2 also contains a single unique sequence but in this case it is flanked by a variable number of repeated sequences at each terminus (e.g. human herpesvirus 8; HHV-8). Structure 3 contains different elements at each terminus, which are present internally in inverted orientation. The genome is thus divided into two unique regions (one “long” and one “short”), which are flanked by inverted repeats. The repeated sequence flanking the long unique sequence is very short (88 bp in human herpesvirus 3; HHV-3) or absent (e.g. suid herpesvirus 1 [SuHV-1] and equid herpesvirus 1 [EHV-1]). Homologous recombination in replicated concatemeric DNA results in inversion of the two regions, and cleavage largely or entirely at one of the two junction regions results in unit length genomes that are one or the other of two isomers differing in the orientation of the short unique sequence. Structure 4 is the most complex. Like structure 3, it contains long and short unique regions, but in this case, both are flanked by large inverted repeat sequences. Homologous recombination and cleavage with equal probability at either of the two junction regions results in the formation of four isomers differing in the orientations of the unique sequences, with each isomer equimolar in virion populations (e.g. HHV-1). In addition, structure 4 genomes contain a short terminal repeat, which is present internally in inverse orientation in the junction region. The different isomers of type 3 and 4 genomes appear to be functionally equivalent.
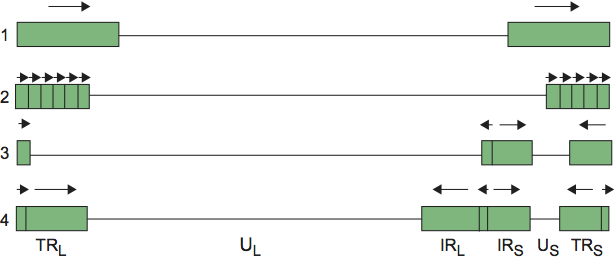
Figure 2. Simplified illustration of herpesvirus genome structures. Unique and repeated sequences are shown as solid lines and rectangles, respectively. The orientations of repeated sequences are indicated by arrows. The nomenclature used to describe regions of type 3 and 4 genomes is shown: UL=unique long; US=unique short. The repeated sequences flanking the unique regions are named “terminal repeat short” (TRS) and “internal repeat short” (IRS), etc.
It should be emphasized that Figure 2 is a simplified depiction of herpesvirus genome structures. Some herpesviruses contain large repetitive elements within the genome that are unrelated to those found at the termini (e.g. human herpesvirus 4; HHV-4), and a more complex set of structures can be cataloged if these are included. Particular genome structures are associated with certain herpesvirus taxa. Thus, the presence of multiple repeated elements at both termini (structure 2) is associated with the subfamily Gammaherpesvirinae (though not all members have this structure) while structure 3 is associated with the genus Varicellovirus. However, distantly related viruses may have equivalent genome structures, which have presumably evolved independently. For example, HHV-6 (family Herpesviridae) and ictalurid herpesvirus 1 (IcHV-1; family Alloherpesviridae) share structure 1, while HHV-1 (family Herpesviridae, subfamily Alphaherpesvirinae) and human herpesvirus 5 (HHV-5; family Herpesviridae, subfamily Betaherpesvirinae) share structure 4.
Proteins
The polypeptide composition of the mature virion varies greatly among herpesviruses. More than 30 different polypeptides have been identified in HHV-1 virions; others likely remain to be found. The mature capsid is composed of four major and several minor proteins, while the tegument contains at least 15 different polypeptides, many of which are dispensable in vitro and are therefore not required for virion morphogenesis. The viral envelope contains at least 10 (and in some viruses many more) integral membrane proteins, a subset of which is required for adsorption and penetration of the host cell. Host proteins can be present in virions, but their functional significance has not been demonstrated.
Lipids
The lipid composition of few herpesvirus envelopes has been reported. The lipid composition of the HHV-1 envelope is reported to resemble that of Golgi membranes more closely than that of other cellular membranes.
Carbohydrates
Virion envelopes contain multiple proteins that carry N-linked and O-linked glycans. Mature, cell-free virions contain complex glycans, whereas a proportion of intracellular virions contain N-linked glycans of the immature high mannose type.
Genome organization and replication
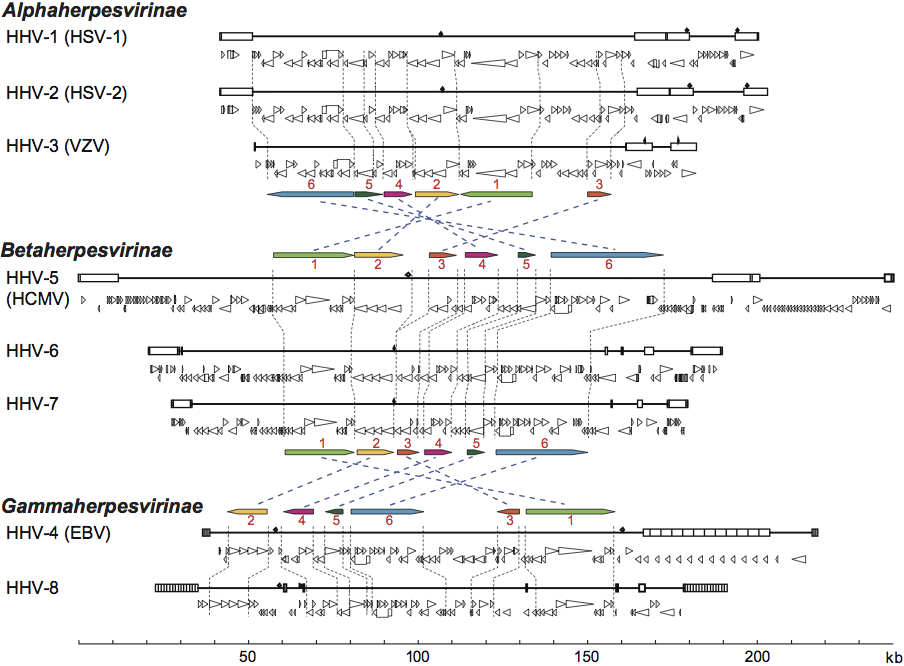
The number of ORFs contained within herpesvirus genomes that potentially encode proteins ranges from about 70 to more than 200. In addition to proteins, herpesvirus genomes also harbor varying numbers of microRNA genes (none yet identified for some viruses and 33 for HHV-4), and express numerous putatively non-translated transcripts of unknown function. A subset of about 40 protein-coding genes is conserved among the viruses of mammals and birds (family Herpesviridae), arranged into six gene blocks (Figure 3). The conserved gene blocks have different orders and orientations in different herpesvirus subfamilies, but genes within a block generally maintain order and transcriptional polarity. The conserved genes encode capsid proteins, components of the DNA replication and packaging machinery, nucleotide modifying enzymes, membrane proteins and tegument proteins, and to a lesser extent control proteins. This reinforces the view that, despite their genetic diversity, these viruses share common features in many aspects of their replication strategies. Members of the three families in the order Herpesvirales are phylogenetically very distant from each other, detectably sharing only two genes (encoding DNA polymerase and the putative ATPase subunit of terminase) derived from a common ancestor, plus a few additional genes that were probably captured independently. The unifying feature across the order Herpesvirales is virion morphology rather than genetic content.
Given the genetic diversity of members of the order Herpesvirales, it is probable that the details of their replication strategy vary, perhaps substantially. What follows, therefore, is a brief description based on well-studied members of the group, HHV-1 in particular (Figure 4). Adsorption and penetration involve the interaction of multiple virion envelope proteins with multiple cell surface receptors. Entry takes place by membrane fusion either at the cell surface or following endocytosis of the attached virion. The nucleocapsid is transported to the region of a nuclear pore by retrograde microtubule transport, while tegument proteins, many of whose functions are unknown, are thought to modify cellular metabolism. For HHV-1, one tegument protein (the UL41 gene product, vhs) acts in the cytoplasm to inhibit host protein synthesis while another (the UL48 gene product, VP16) is a transcription factor that enters the nucleus and activates viral immediate early genes. In permissive cells, entry of the genome into the nucleus is followed by a transcriptional cascade. Immediate early (α) genes, which are largely distinct among the various families and subfamilies, regulate subsequent gene expression by transcriptional and post-transcriptional mechanisms. Early (β) genes encode the DNA replication complex and a variety of enzymes and proteins involved in modifying host cell metabolism, while the structural proteins of the virus are encoded primarily by late (γ) genes. Immediate early genes can be transcribed in the absence of de novo protein synthesis. Early gene transcription is dependent on expression of immediate early proteins. Late gene transcription is dependent on viral DNA synthesis. With the exception of a small number of non-translated RNAs expressed by specific herpesviruses, transcription is by host RNA polymerase II.
 \
\
Figure 3. Genomic and genetic architectures of the human herpesviruses. Major repeat elements are indicated on each genomic schematic as boxes. Beneath each genome, ORFs considered likely to encode expressed proteins are indicated as triangles that are oriented to show their direction of transcription. 5′-exons of spliced genes are indicated as boxes that are connected by bars to 3′-exons. The six conserved herpesvirus sequence blocks (Block 1 through Block 6) are diagrammed to show their relative locations and orientations in the three major lineages. Diagrams are based on coordinates in GenBank accession numbers NC_001806 (HSV-1 strain 17), NC_001798 (HSV-2 strain HG52), NC_001348 (VZV strain Dumas), NC_001347 (HCMV), NC_000898 (HHV-6B strain Z29), U43400 and NC_001716 (HHV-7 strains JI and RK), NC_007605 (EBV strain B95-8) and U75698 (HHV-8, strain BC-1). (Adapted from Pellett, P.E. and Roizman, B. (2007). The Herpesviridae: a brief introduction In: Fields Virology, 5th edn (D.M. Knipe and P.M. Howley, Eds.), Lippincott, Williams, & Wilkins, Philadelphia, vol. 2, ch. 66, pp. 2479–2499; with permission of Lippincott Williams & Wilkins.)
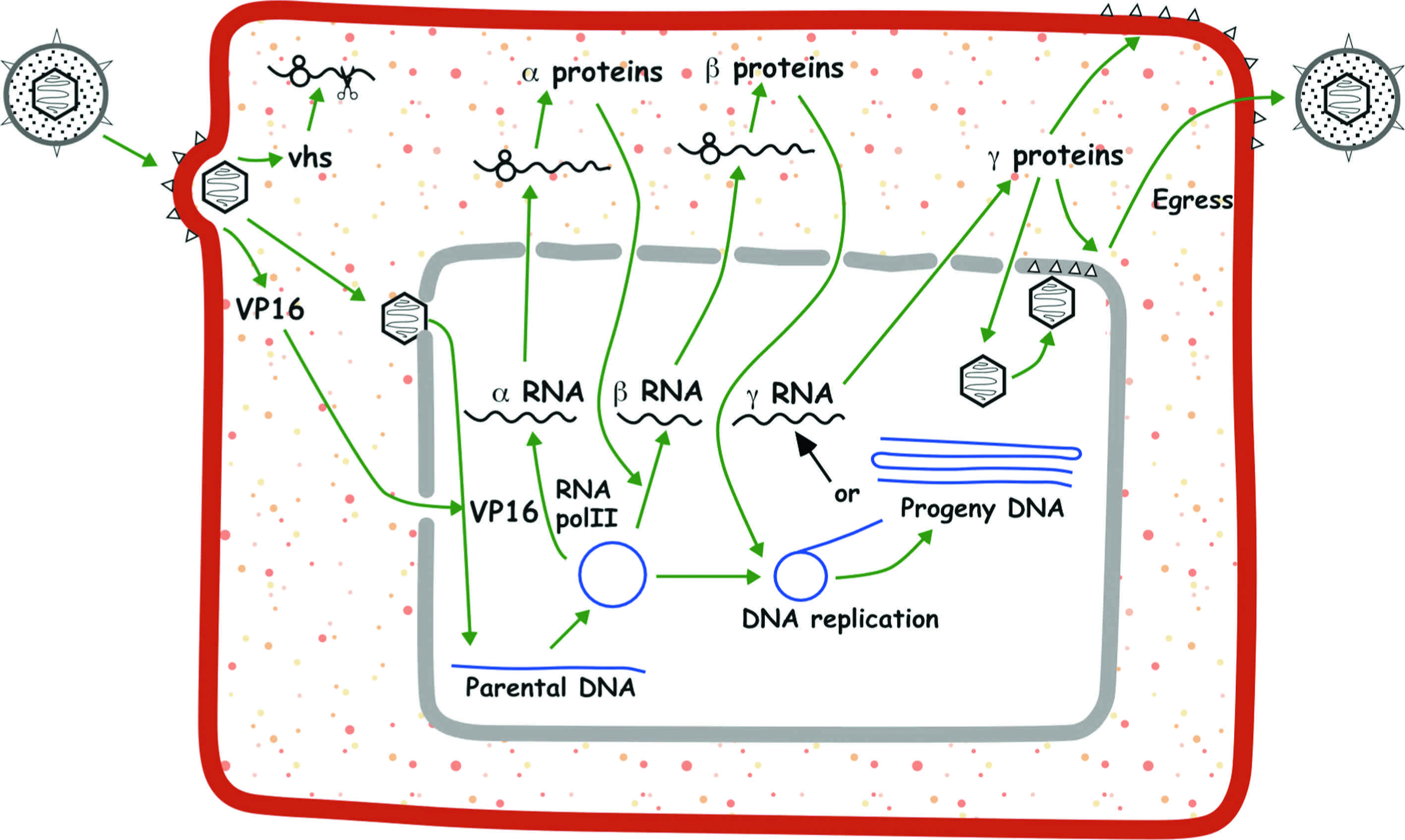
Figure 4. Schematic representation of the lytic replication cycle of human herpesvirus 1 (HHV-1) in permissive cells. (From Roizman, B. and Knipe, D.M. (2001). Herpes simplex viruses and their replication. In: Fields Virology, 4th edn (D.M. Knipe and P.M. Howley, Eds.), Lippincott Williams & Wilkins, Philadelphia, vol. 2, ch. 72, pp. 2399–2459; with permission of Lippincott Williams & Wilkins.)
Viral DNA synthesis occurs from one or more origins of replication, probably by a rolling circle mechanism. In HHV-1, DNA replication requires seven gene products: an origin-binding protein, an ssDNA-binding protein, a DNA polymerase composed of two subunits and a helicase-primase complex composed of three gene products. Homologs of all but the origin-binding protein have been identified in members of all three subfamilies in the family Herpesviridae. Newly synthesized DNA is packaged from the concatemer into preformed immature capsids within the nucleus by processes that involve several viral proteins. Immature capsids contain a core of scaffolding proteins, which are expelled after proteolytic cleavage during maturation. The subsequent steps in morphogenesis of secreted enveloped virions are a subject of ongoing experimentation and debate. Nucleocapsids are observed budding through the inner nuclear membrane into the perinuclear space. One line of evidence favors the view that the enveloped particles in the perinuclear space become “de-enveloped” by fusion with the outer nuclear membrane and that the resulting nucleocapsids are re-enveloped in a Golgi or post-Golgi compartment. Alternatives include (i) envelopment at the inner nuclear membrane and subsequent transit to the plasma membrane in transport vesicles, and (ii) nuclear egress via dilated nuclear pore complexes, followed by envelopment at a cytoplasmic vesicle that is transported to the plasma membrane for virion release. Only a subset of herpesvirus genes is required to achieve this basic replication cycle in vitro. Almost half the genes of HHV-1 are not individually required for replication in cultured cells; the products of these auxiliary genes (sometimes described as being non-essential for replication in cell culture) have diverse and often significant functions in vivo.
The alternative to the productive cycle, and consequent cell death, is latent infection. The establishment and maintenance of the latent state is not thoroughly understood, but the weight of evidence favors a “default” mechanism in which failure of immediate early gene expression leads to maintenance of the input genome as a circular episomal element. It has been reported that some latently-infected neurons contain large numbers of HHV-1 DNA copies, suggesting that a latent state can be established after initiation of the productive cycle. Changes in the transcription factor milieu of the latently infected cell, due to external stimuli or cell differentiation, lead to immediate early gene expression and entry into the productive cycle.
Like other large eukaryotic DNA viruses, herpesviruses are used as vectors for gene therapy.
Antigenic properties
Infected hosts produce antibodies and cell-mediated immune responses to many structural and non-structural virus proteins. Individual antigenic proteins can harbor multiple epitopes to which antibody and cell mediated responses are elicited. Some of the envelope glycoproteins are particularly immunogenic and are targets for neutralizing antibodies. Efficient cross-neutralization is observed only between closely related viruses within a genus.
Biological properties
The range of host species is very wide. It is probable that all vertebrates carry multiple herpesvirus species, and a herpesvirus has also been identified in invertebrates (molluscs). As a general rule, the natural host range of individual viruses is highly restricted, and most herpesviruses are thought to have evolved in association with single host species. However, there are many examples of cross-species transmission that sometimes result in severe disease and even death. This can lead to misidentification of the natural host species. Cross-species transmission is most likely among related hosts, e.g. among equids. SuHV-1 has an exceptional and remarkably wide host range in the wild, causing fatal disease in unrelated species following natural modes of transmission.
Host range varies considerably in experimental animal systems: some members of the subfamily Alphaherpesvirinae can infect a wide variety of animal species, whereas members of the subfamilies Betaherpesvirinae and Gammaherpesvirinae exhibit a very restricted experimental animal host range. Host range in vitro also varies considerably, though the same general rule holds true: members of the subfamily Alphaherpesvirinae will often infect a variety of cells of diverse mammalian species in vitro, whereas members of the subfamilies Betaherpesvirinae and Gammaherpesvirinae exhibit greater restriction. The basis of host restriction both in vivo and in vitro is poorly understood. In a few instances (e.g. HHV-4), cell surface receptors play an important part in determining host range, but more commonly the virion is capable of entering a wide variety of cells, with intracellular factors determining susceptibility (e.g. HHV-5). Natural transmission routes range from highly contagious aerosol spread (HHV-3, equid herpevirus 4 [EHV-4] and felid herpesvirus 1 [FHV-1]) to intimate oral contact (HHV-4) or sexual contact (human herpesvirus 2 [HHV-2]); HHV-6 can be transmitted rarely via the host germ line as an integrated viral genome. Unusually, the alphaherpesvirus gallid herpesvirus 2 (GaHV-2) is shed predominantly from the base of chicken feather follicles and transmitted as an airborne, inhaled dander. Vector-mediated transmission has not been reported.
Herpesviruses are highly adapted to their hosts, and severe infection is usually observed only in the very young, the fetus, the immunocompromised, or following infection of an alternative host. Most herpesviruses establish a systemic infection, a cell-associated viraemia being detectable during primary infection. Some members of the genus Simplexvirus appear to be an exception to the rule: in the normal host, infection is limited to epithelium at the site of infection and sensory nerves innervating the site. A variety of immune evasion mechanisms have been identified in different viruses, including those operating against complement, antibody, MHC class I presentation and NK cell killing. The key to survival of herpesviruses is their ability to establish life-long latent infection, a feature that is assumed to be the hallmark of all herpesviruses. The cell type responsible for harboring latent virus has been established in relatively few instances. Nevertheless, the emerging picture is that for the family Herpesviridae, members of the subfamily Alphaherpesvirinae establish latent infection in neurons, members of the subfamily Betaherpesvirinae establish latent infection in cells of the monocyte series, and members of the subfamily Gammaherpesvirinae establish latent infection in lymphocytes. It should be emphasized, however, that this general picture is based on a very limited number of examples and that there are reports of latent infection at other sites. Little detail is known about latency for the families Alloherpesviridae and Malacoherpesviridae.
Phylogenetic relationships within the order
Members of the order Herpesvirales comprise a diverse collection of viruses that share characteristic virion morphology. The protein that comes nearest to being herpesvirus-specific is the putative ATPase subunit of the terminase (a complex that is responsible for packaging viral DNA into nascent capsids), which is conserved in all herpesviruses and has more distant relatives in T4-like bacteriophages of the family Myoviridae. The diversity of the order has meant that criteria such as serology or nucleic acid hybridization have limited value in determining relationships between different viruses, and the construction of a satisfactory taxonomic structure has been a significant challenge. Historically, members of the family Herpesviridae were grouped into subfamilies on the basis of broad biological criteria. Division of the subfamilies into genera was based on antigenic cross-reactivity and molecular criteria, primarily the size and structure of the genome. In retrospect, these assignments were found to be generally consistent with sequence-based phylogeny. Modern classification of herpesviruses is based primarily on genetic content.
At all levels from order to species, herpesvirus taxa are described as corresponding to “distinct genetic lineages”; these lineages are defined by two criteria: (a) comparison of nucleotide or predicted amino acid sequences of conserved herpesvirus genes, and (b) identification of particular genes or genetic properties that are unique to a virus subset.
Phylogenetic relationships within the family Herpesviridae are illustrated in Figure 5.
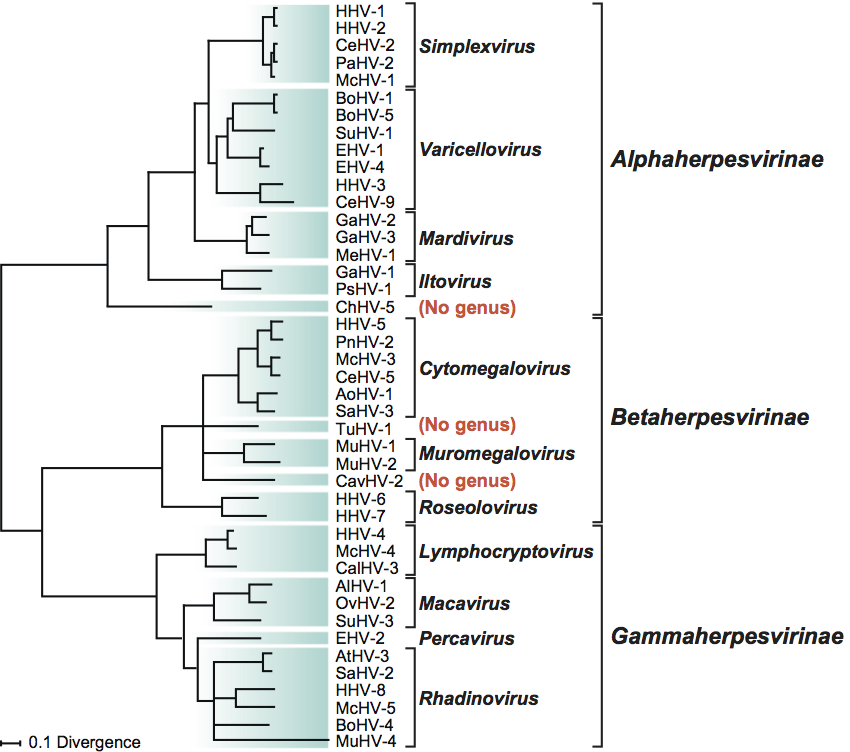
Figure 5. Phylogenetic relationships within the family Herpesviridae. The unrooted Bayesian tree is based on amino acid sequence alignments for the orthologs of HHV-1 genes UL15, UL19, UL27, UL28, UL29 and UL30. The scale indicates the number of amino acid substitutions per site. (Adapted from McGeoch, D.J., Davison, A.J., Dolan, A., Gatherer, D. and Sevilla-Reyes, E.E. (2008). Molecular evolution of the Herpesvirales. In: Origin and Evolution of Viruses, 2nd edn (E. Domingo, C.R. Parrish and J.J. Holland, Eds.), Elsevier, London, pp. 447–475; used with permission from Elsevier.)
Species demarcation criteria within the order
A herpesvirus may be classified as a species if it has distinct epidemiological or biological characteristics and a distinct genome that represents an independent replicating lineage.
Sequence information is required for formal recognition of new herpesvirus species. Replicating lineages of herpesviruses are now identified primarily on the basis of information derived from genomic sequences. Sequence information sufficient to demonstrate that a novel virus represents a replicating lineage distinct from known herpesvirus species is taken as evidence that the virus in question exists in nature, occupies a distinct ecological niche and thus can be recognized as a herpesvirus species. For some well-studied genes, there are levels of sequence difference beyond which there are no instances in which the viruses in question do not have distinct epidemiological and biological properties; such viruses can be reliably recognized as species on the basis of limited sequence information. There are also closely related viruses that have relatively small differences in the sequences of individual genes, but genetic differences extend across the respective genomes in a manner indicative of them representing independent replicating lineages. These viruses also have distinct epidemiological and biological characteristics (e.g. host identity, pathogenic and epidemiological properties, and the lack of occurrence of natural recombinants) and thus meet the definition of herpesvirus species.
Similarity with other taxa
Herpesviruses possess several genes (e.g. encoding enzymes or immunomodulatory factors) that are related to cellular genes and are assumed to have been gained by capture. The equivalent feature in certain other virus families results in genetic similarities that probably indicate independent capture events rather than direct evolutionary relationships. One speculative exception is the putative ATPase subunit of the DNA packaging terminase complex in T4 and related dsDNA bacteriophages, which has distant counterparts in all herpesviruses.
Nomenclature of herpesvirus species
A herpesvirus species name consists of three parts.
- A term derived from a taxon of the host that in its natural setting harbors the virus. The default taxon employed is that of family, and, except for the species of humans, it ends in ‘-id’. Exceptions are species from the family Bovidae, which are designated by host subfamily or genus, and nonhuman primates (host genus); these names end in ‘-ine’.
- The word “herpesvirus”.
- An Arabic numeral, which, in combination with (i), provides a unique name.
Derivation of names
Allo: from Greek allo, “other, different”.
Alpha: Greek α, “a”.
Batracho: from Greek batrachos, “frog”.
Beta: Greek β, “b”.
Cyprini: from Latin cyprinus, “carp”.
Cytomegalo: from Greek kytos, “cell”, and megas, “large”.
Gamma: Greek γ, “g”.
Herpes: from Greek herpes, “creeping”.
Ictaluri: from Ictaluridae.
Ilto: from infectious laryngotracheitis
Lymphocrypto: from Latin lympha, “water”, and Greek kryptos, “concealed”.
Maca: from malignant catarrhal fever.
Malaco: from Greek malaco, “soft”, as applied to molluscs.
Mardi: from Marek’s disease.
Muromegalo: from Latin mus, “mouse”, and Greek megas, “great”.
Ostrea: from Greek ostreo or Latin ostrea, “oyster”.
Perca: from perissodactyl and carnivore.
Probosci: from Greek and Latin proboscis, “elephant’s trunk”.
Rhadino: from Greek rhadinos, “slender, taper”.
Roseolo: from Latin rose, “rose, rosy”.
Simplex: from Latin simplex, “simple”.
Varicello: from Latin varius, “spotted”, and its diminutive variola, “smallpox”.
Further reading
Chiu, W. and Rixon, F.J. (2002). High resolution structural studies of complex icosahedral viruses: a brief overview. Virus Res., 82, 9-17.
Davison, A.J. (2007). Introduction: definition and classification of the human herpesviruses, comparative analysis of the genomes. In: A. Arvin, G. Campidelli-Fiume, E. Mocarski, P.S. Moore, B. Roizman, R. Whitley and K. Yamanishi (Eds.), Human Herpesviruses: Biology, Therapy and Immunoprophylaxis. Cambridge University Press, Cambridge, pp. 10-26.
Davison, A.J. (2010). Herpesvirus systematics. Veterin. Microbiol., 143, 52-69.
Davison, A.J., Eberle, R., Ehlers, B., Hayward, G.S., McGeoch, D.J., Minson, A.C., Pellett, P.E., Roizman, B., Studdert, M.J. and Thiry, E. (2009). The order Herpesvirales. Arch. Virol., 154, 171-177.
McGeoch, D.J., Rixon, F.J. and Davison, A.J. (2006). Topics in herpesvirus genomics and evolution. Virus Res., 117, 90-104.
Mettenleiter, T.C., Klupp, B.G. and Granzow, H. (2009). Herpesvirus assembly: an update. Virus Res., 143, 222-234.
Pellett, P.E. and Roizman, B. (2007). The Family Herpesviridae: a brief introduction. In: D.M. Knipe, P.M. Howley, D.E. Griffin, R.A. Lamb, M.A. Martin, B. Roizman, S.E. Straus (Eds), Fields Virology, 5th edn. Lippincott Williams and Wilkins, Philadelphia, PA, pp. 2479-2499.
Roizman, B., Whitley, R.J. and Knipe, D.M. (2007). Herpes simplex viruses. In: D.M. Knipe, P.M. Howley, D.E. Griffin, R.A. Lamb, M.A. Martin, B. Roizman and S.E. Straus (Eds), Fields Virology, 5th edn. Lippincott Williams and Wilkins, Philadelphia, PA, pp. 2501-2601.
Contributed by
Pellett, P.E. (Chair), Davison, A.J., Eberle, R., Ehlers, B., Hayward, G.S., Lacoste, V., Minson, A.C., Nicholas, J., Roizman, B., Studdert, M.J. and Wang, F.
