Family: Fuselloviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Fusellovirus
Type species Sulfolobus spindle-shaped virus 1
Virion properties
Morphology
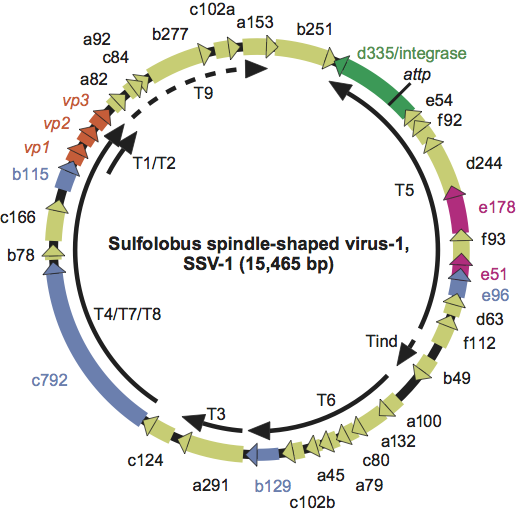
Virions are lemon-shaped, with short tail fibers attached to one pole, and slightly heterogeneous in size. Virions are 55–60 nm in their short dimension and 80–100 nm in their long dimension. A small fraction of the Sulfolobus spindle-shaped virus 1 (SSV-1) population (up to 1%) is larger, with a particle length of about 300 nm. Some other fuselloviruses, particularly SSV-K1, have more elongated virions. Purified SSV-1 virions contain host lipids and three virus encoded proteins, two of which are associated with the coat and the other is DNA-associated.
Physicochemical and physical properties
Virion buoyant density in CsCl is 1.24 g cm−3. The particles are stable at up to 97 °C and are insensitive to urea, ether and pH 2. However, low pH (below 5) reduces viability due to degradation of the DNA, and virions are sensitive to pH above 11 and trichloromethane.
Nucleic acid
Virions contain circular dsDNA, from 14.8 to 17.8 kbp. In SSV1 virions, DNA is positively supercoiled and associated with polyamines and a virus-coded basic protein.
Negative contrast electron micrograph of Sulfolobus spindle-shaped virus 1 (SSV-1) virions, bound to a membrane vesicle (left) or isolated (right) (Stedman et al., 1999). The bars represent 100 nm.
Proteins
The main constituents of the SSV-1 viral envelope are two basic proteins (VP1 and VP3). In SSV-1, these proteins are 73 and 92 aa, respectively, as deduced from the DNA sequence and from N-terminal protein sequencing. The VP1 protein is post-translationally proteolytically processed. In SSV-1, a very basic protein (VP2, 74 aa) is attached to the viral DNA. However, this gene is lacking in all other sequenced fusellovirus genomes. The genes encoding these structural proteins are closely linked in the fusellovirus genomes, in the order VP1, VP3 and VP2 in SSV-1 (Figure 2). The second largest ORF of SSV-1 (ORF d335, 335 aa) is similar to the integrase family of site-specific tyrosine recombinases. This protein has been expressed in E. coli and recombines DNA fragments sequence-specifically in vitro. The gene is conserved in all sequenced fuselloviruses. Four ORFs in the virus genome have been shown to be essential for virus function and two have been shown to be non-essential for virus function in SSV-1 (see Figure 2).
Lipids
It has been reported that 10% of the SSV-1 virion envelope consists of host lipids.
Carbohydrates
None reported.
Genome organization and replication
The virus genome is present in cells as circular dsDNA and is site-specifically integrated into a tRNA gene of the host chromosome. In SSV-1 the integrated copy is flanked by a 44 bp direct repeat (attachment core) that occurs once in the SSV-1 circular DNA genome. Upon integration, the viral integrase gene is disrupted. Eleven somewhat overlapping transcripts originating from seven promoters have been detected and mapped. They almost completely cover the SSV-1 genome (Figure 2). UV-irradiation stimulates virus production and progeny virions are released without host cells lysis. A small transcript (Tind) is strongly induced upon ultraviolet induction. Particles appear to be assembled and are produced by extrusion at the cell membrane.
Antigenic properties
Not known.
Biological properties
The host range of the fuselloviruses is limited to extremely thermophilic Archaea: Sulfolobus shibatae, Sulfolobus solfataricus strains P1 and P2 and Sulfolobus islandicus strains. Very few phage particles are produced in cultures of SSV-1 lysogens. UV-irradiation strongly induces SSV-1 production without evident lysis of the host. Fuselloviruses have been found in about 8% of Sulfolobus isolates from Icelandic solfataric fields. They have also been found in solfataric hot springs in Yellowstone National Park in the USA and on the Kamchatka peninsula in the Russian Federation. An infectious shuttle vector that also replicates in E. coli has been constructed.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Fusellovirus
| Sulfolobus spindle-shaped virus 1 | ||
| Sulfolobus spindle-shaped virus 1 | ||
| Sulfolobus shibatae | [X07234] | (SSV-1-Ss) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Fusellovirus but have not been approved as species
| Sulfolobus spindle-shaped virus 2 | [AY370762] | (SSV-2) |
| Sulfolobus spindle-shaped virus 3 |
| (SSV-3) |
| Sulfolobus spindle-shaped virus 4 | [EU030938] | (SSV-4) |
| Sulfolobus spindle-s haped virus 5 | [EU030939] | (SSV-5) |
| Sulfolobus spindle-shaped virus 6 | [FJ870915] | (SSV-6) |
| Sulfolobus spindle-shaped virus 7 | [FJ870916] | (SSV-7) |
| Sulfolobus spindle-shaped virus - Kamchatka 1 | [AY423772] | (SSV-K1) |
| Sulfolobus spindle-shaped virus - Yellowstone 1 | [AY388628] | (SSV-Y1) |
| Acidianus spindle-shaped virus 1 | [FJ870917] | (ASV-1) |
List of unassigned species in the family Fuselloviridae
None reported.
Phylogenetic relationships within the family
Phylogenetic relationships within the family Fuselloviridae are unclear. Different homologous genes give different phylogenetic trees.
Similarity with other taxa
One ORF in fusellovirus genomes is similar to a gene present in the Sulfolobus viruses of the families Lipothrixviridae and Rudiviridae. The virus of extreme halophiles, His1, originally suggested to be a fusellovirus, on further characterization has been found to be very dissimilar and a novel genus (Salterprovirus) has been established for this group of viruses.
Derivation of name
Fusello: from the Latin fusello, “little spindle”.
Further reading
Palm, P., Schleper, C., Grampp, B., Yeats, S., McWilliam, P., Reiter, W.-D. and Zillig, W. (1991). Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology, 185, 242-250
Peng, X. (2008). Evidence for the horizontal transfer of an integrase gene from a fusellovirus to a pRN-like plasmid within a single strain of Sulfolobus and its implications for plasmid survival. Microbiol., 154, 383-391.
Prangishvili, D., Stedman, K. and Zillig, W. (2001). Viruses of the extremely thermophilic archaeon Sulfolobus. Trends Microbiol., 9, 39-43.
Redder, P., Peng, X, Brügger, K., Shah, S.A., Roesch, F., Greve, B., She, Q., Schleper, C., Forterre, P., Garrett, R. A. and Prangishvili, D. (2009). Four newly isolated fuselloviruses from extreme geothermal environments reveal unusual morphologies and a possible inter-viral recombination mechanism. Environm. Microbiol., 11, 2849–2862.
Stedman, K.M., Schleper, C., Rumpf, E. and Zillig, W. (1999). Genetic requirements for the function of the archaeal virus SSV1 in Sulfolobus solfataricus: Construction and testing of viral shuttle vectors. Genetics, 152, 1397-1406.
Stedman, K.M., She, Q., Phan, H., Arnold, H.P., Holz, I., Garrett, R.A. and Zillig, W. (2003). Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV-1 and SSV-2. Res. Microbiol., 154, 295-302.
Zillig, W., Prangishvili, D., Schleper, C., Elferink, M., Holz, I., Albers, S., Janekovic, D. and Götz, D. (1996). Viruses, plasmids and other genetic elements of thermophilic and hyperthermophilic Archaea. FEMS Microbiol. Rev., 18, 225-236.
Zillig, W., Reiter, W.D., Palm, P., Gropp, F., Neumann, H. and Rettenberger, M. (1988). Viruses of archaebacteria. In: R. Calendar (Ed.), The Bacteriophages, vol 1. Plenum Press, New York, pp. 517-558.
Contributed by
Prangishvili, D.
The author acknowledges the contribution to the Eighth ICTV Report of Stedman, K.M.
Figures
Figure 1 Negative contrast electron micrograph of Sulfolobus spindle-shaped virus 1 (SSV-1) virions, bound to a membrane vesicle (left) or isolated (right) (Stedman et al., 1999). The bars represent 100 nm.
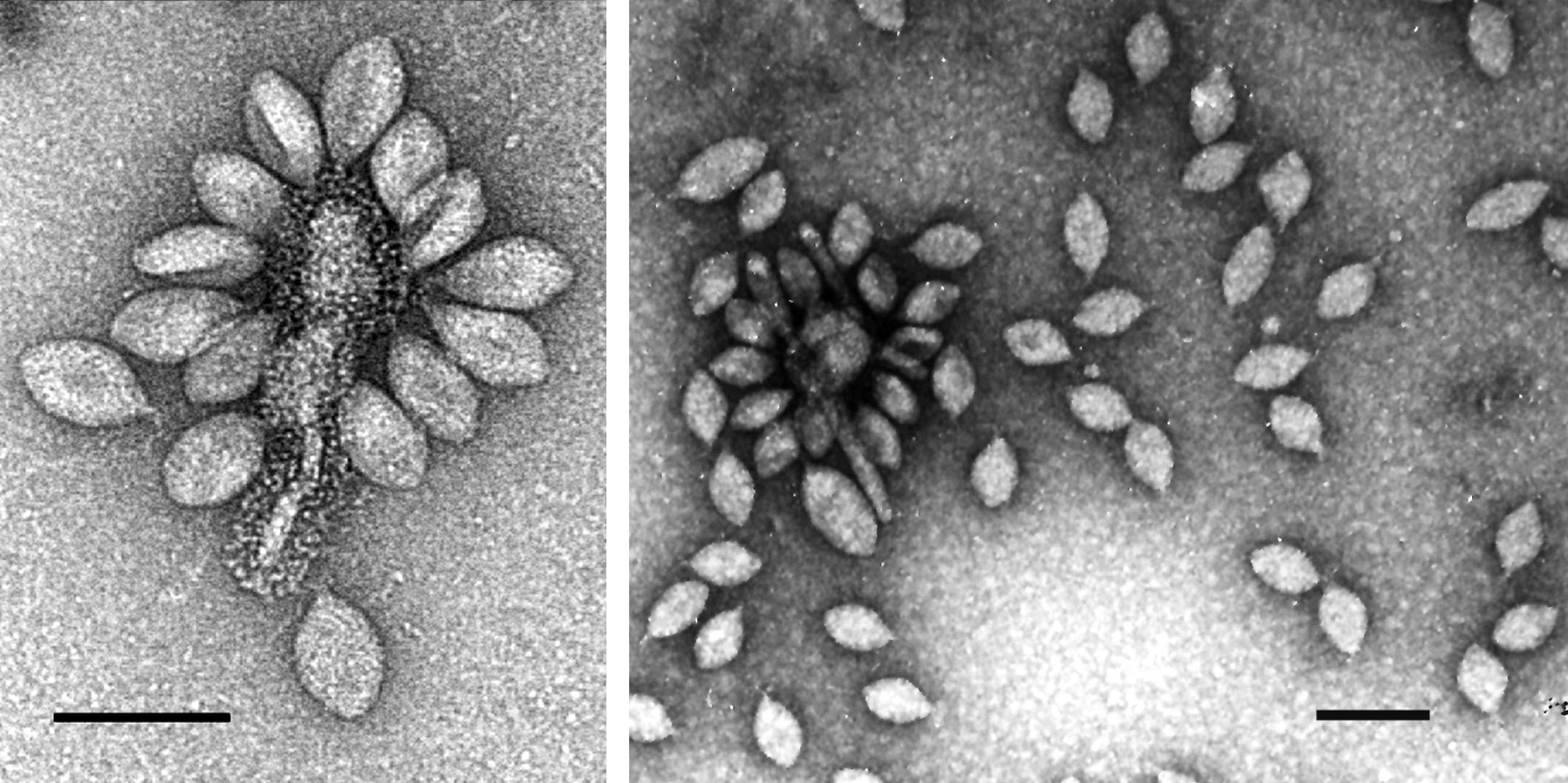
Figure 2 Genome organization of Sulfolobus spindle-shaped virus 1 (SSV-1). ORFs are shown as arrows and labeled with gene names. The major virion protein genes (VP1, VP3 and VP2) are labeled in red. The viral integrase gene, ORF d335, is labeled in green. ORFs shown to be essential for virus function are labeled in blue. ORFs shown to be non-essential for virus function are labeled in pink. The viral attachment site is labeled as attP. Transcripts are shown as solid arrows and labeled. Transcripts T1 and T2 start at the same promoter and overlap, and transcripts T4, T7 and T8 similarly overlap. The termination site of transcript T9 is not known.