Order Caudovirales
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
The order consists of the three families of tailed bacterial viruses infecting Bacteria and Archaea: Myoviridae (long contractile tails), Siphoviridae (long non-contractile tails) and Podoviridae (short non-contractile tails). Tailed bacterial viruses are an extremely large group with highly diverse virion, genome and replication properties. Over 4500 descriptions have been published (accounting for 96% of reported bacterial viruses): 24% in the family Myoviridae, 62% in the family Siphoviridae and 14% in the family Podoviridae (as of November 2001). However, data on virion structure, genome organization and replication properties are available for only a small number of well-studied species. Although extensive horizontal gene transfer between bacterial cells and viruses has obscured phylogenetic relationships amongst some tailed viruses, particularly those which are temperate, enough common features still survive to indicate their fundamental relatedness. Therefore, formal taxonomic names are used for Caudovirales at the order and family level, but only vernacular names at the genus level. Since publication of the Eighth Report, some taxonomic revision of the family Podoviridae has been accomplished based on genome information. This has led to the introduction of subfamilies to accommodate the wealth and diversity of bacterial viruses being discovered.
Virion properties
Morphology
The virion has no envelope and consists of two parts, the head and the tail. The head is a protein shell and contains a single linear dsDNA molecule, and the tail is a protein tube whose distal end binds the surface receptors on susceptible bacterial cells. DNA travels through the tail tube during delivery (often called “injection”) into the cell being infected. Heads have icosahedral symmetry or elongated derivatives thereof (with known triangulation numbers of T=4, 7, 13, 16 and 52). Capsomers are seldom visible: heads usually appear smooth and thin-walled (2–3 nm). When they are visible, morphological features (capsomeres) on the surface of the head commonly form 72 capsomers (T=7; 420 protein subunits), but known capsomer numbers vary from 42 to 522. Isometric heads are typically 45–170 nm in diameter. Elongated heads derive from icosahedra by addition of equatorial belts of capsomers and can be up to 230 nm long. DNA forms a tightly packed coil (without bound proteins) inside the head. Tail shafts have six-fold or (rarely) three-fold symmetry, and are helical or stacks of disks of subunits from 3 and 825 nm in length. They usually have base plates, spikes, or terminal fibers at the distal end. Some viruses have collars at the head–tail junction, head or collar appendages, transverse tail disks, or other attachments.
Physicochemical and physical properties
Virion Mr is 20–600 ×106; S20,w values are 200 to >1200S. Both upper limits may be underestimates, since these properties have not been determined for the largest tailed viruses. Buoyant density in CsCl is typically about 1.5 g cm−3. Most tailed viruses are stable at pH 5–9; a few are stable at pH 2 or pH 11. Heat sensitivity is variable, but many virions are inactivated by heating at 55–75 °C for 30 min. Tailed viruses are rather resistant to UV irradiation. Heat and UV inactivation generally follow first-order kinetics. Most tailed phages are stable to chloroform. Inactivation by nonionic detergents is variable and concentration-dependent. Some virions are sensitive to osmotic shock, and many are sensitive to Mg+ chelators.
Nucleic acid
Virions contain one molecule of linear dsDNA. Genome sizes are 18 to >500 kbp, corresponding to Mr values of 11 to >300 ×106. DNA content is 45–55% of the virions. G+C contents are 27–72% and usually resemble those of host DNA. Some viral DNAs contain modified nucleotides which partially or completely replace normal nucleotides (e.g., 5-hydroxymethylcytosine instead of cytosine), and/or are glycosylated or otherwise modified.
Proteins
There are 7–49 different virion structural proteins. Typical head shells are made up of 60T molecules of a single main building block CP and 12 molecules of portal protein through which DNA enters and leaves, but they can also contain varied numbers of proteins that plug the portal hole, proteins to which tails bind, proteins that bind to the outside of the CP shell (decoration proteins) and other proteins whose roles are not known. Non-contractile tails are made of one major shaft or tube protein and contractile tails have a second major protein, the sheath protein that forms a cylinder around the central tube. Tails also have small numbers of varied specific proteins at both ends. Those at the end distal from the head form a structure called the tail tip (Siphovirus) or baseplate (Myovirus) to which the tail fibers are attached. The tail fibers bind to the first-contact receptors on the surface of susceptible cells. Fibers or baseplates may include proteins with endoglycosidase or peptidoglycan hydrolase activity that aid in gaining access to the cell surface and entry of DNA into the cell. Most virions carry proteins that are injected with the DNA, such as transcription factors, RNA polymerase and others with poorly understood functions.
Lipids
No well-characterized virions contain lipid.
Carbohydrates
Glycoproteins, glycolipids, hexosamine and a polysaccharide have been reported in certain virions but these are not well-characterized.
Genome organization and replication
Genome organization
The linear dsDNA genomes encode from 27 to over 600 genes that are highly clustered according to function and tend to be arranged in large operons. Complete functional genomic maps are very diverse and available for only a relatively small number of tailed viruses. Virion DNAs may be circularly permuted and/or terminally redundant, have single stranded gaps, or have covalently-bound terminal proteins. The ends of these linear molecules can be blunt or have complementary protruding 5′ or 3′ ends (the “cohesive” or “sticky” ends, which can base-pair to circularize the molecule). Prophages of temperate tailed viruses are either integrated into the host genome or replicate as circular or linear plasmids; these linear plasmids have covalently-closed hairpin telomeres.
Replication
In typical lytic infections, after entering the host cell, viral DNA may either circularize or remain linear. A few viruses use terminal proteins to prime DNA replication and package progeny viral DNA (29 and its relatives) or replicate DNA by a duplicative transposition mechanism (Mu and its relatives). Gene expression is largely time-ordered and groups of genes are sequentially expressed. “Early genes” are expressed first and are largely involved in host cell modification and viral DNA replication. “Late genes” specify virion structural proteins and lysis proteins (Figure 1). The larger tailed viruses have gene expression cascades that are more complex than this simple scenario. Transcription often requires host RNA polymerase, but many tailed viruses encode RNA polymerases or transcription factors that affect the host RNA polymerase. Translational control is poorly understood and no generalizations are possible at the present state of knowledge. All tailed viruses encode proteins that direct the replication apparatus to the replication origin, but this apparatus may be entirely host-derived, partly virus encoded, or entirely virus encoded. DNA replication is semi-conservative, may be either bidirectional or unidirectional, and usually results in the formation of concatemers (multiple genomes joined head-to-tail) by recombination between phage DNAs or by rolling circle replication. Progeny viral DNA is generated during virion assembly by cleavage from this concatemeric DNA: (i) at unique sites to produce identical DNA molecules with either cos sites or blunt-ended, terminally redundant termini, (ii) at pac sites to produce circularly permuted, terminal redundant DNAs, or (iii) by a headful mechanism to produce terminally redundant, circularly permuted DNAs.
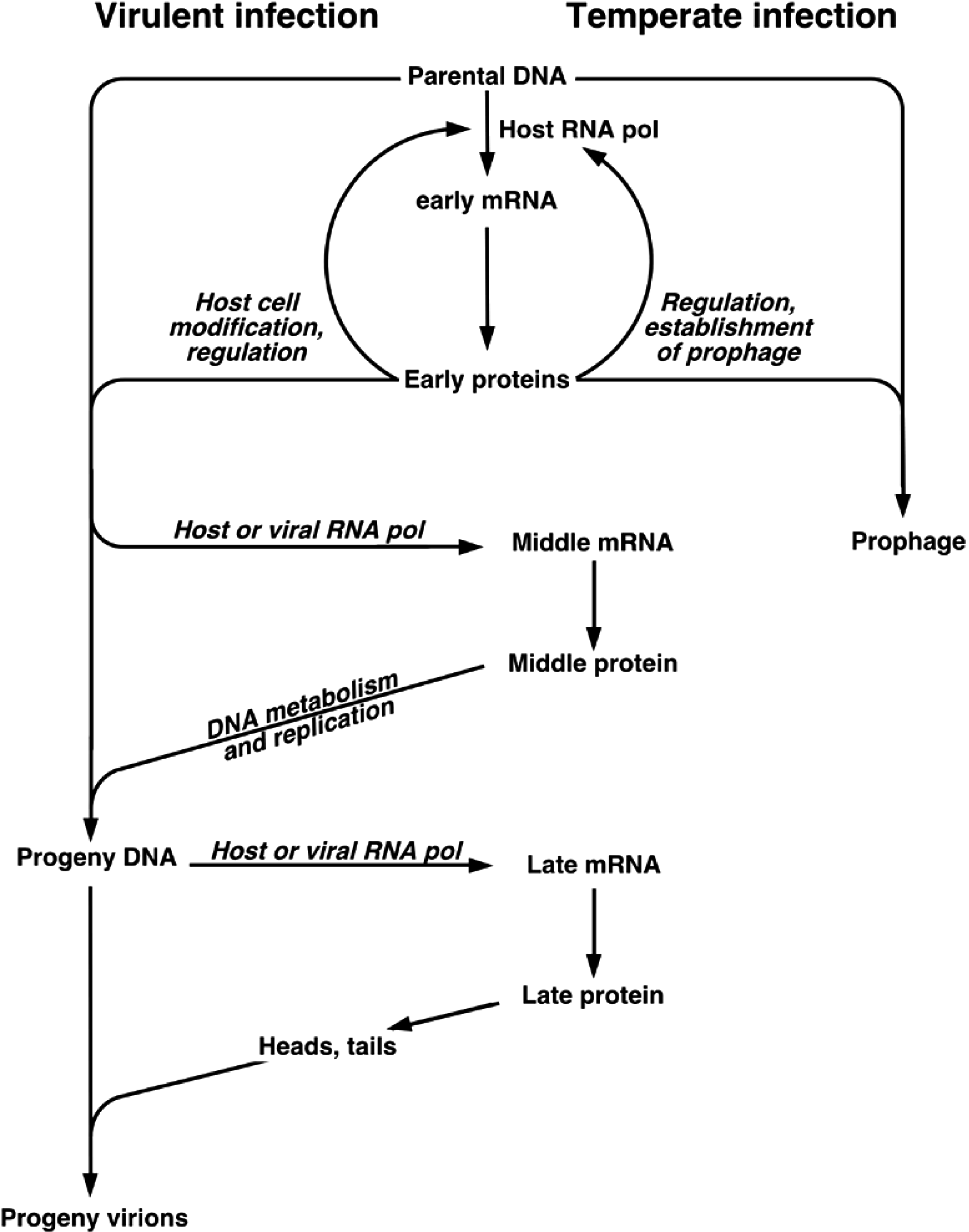 _
_
Figure 1. Flow chart of tailed phage replication. The chart depicts the replication of “typical” virulent phages such as Enterobacteria phage T4 (T4), Enterobacteria phage T7 (T7), and the temperate phages.
Virion assembly and DNA packaging
Assembly of virions from newly made proteins and replicated DNA is complex and generally includes separate pathways for heads, tails and tail fibers. Coat protein shells, called procapsids or proheads, are assembled first, and DNA is inserted into these preformed proteinaceous containers. Assembly of procapsids is poorly understood, but often utilizes an internal scaffolding protein which helps CP assemble correctly and is then released from the shell after its construction. In many, but not all, tailed viruses, proteolytic cleavages (by host or virus-encoded proteases) of some proteins accompany assembly. Virus-specific DNA is recognized for packaging into procapsids by the terminase protein. One end of the DNA is then threaded through the procapsid’s portal structure, and DNA is pumped into the head by an ATP hydrolysis-driven motor that is probably made up of the two terminase subunits and portal protein. Unless unit length DNA molecules are the substrate for packaging (such as with Phi29), when the head is full of DNA a “headful sensing device” recognizes this fact and causes the terminase to cleave the DNA to release the full head from the unpackaged remainder of the DNA concatemer. The terminase subunits are usually released from the virion after DNA is packaged. Filled heads then join to tails and tail fibers to form progeny virions. Some viruses form intracellular arrays, and many produce aberrant structures (polyheads, polytails, giant, multi-tailed, or misshapen particles). Progeny viruses are liberated by lysis of the host cell. Cell lysis is caused by phage-encoded peptidoglycan hydrolases; but lysis timing is controlled by holins, phage encoded inner membrane proteins that allow the hydrolases to escape from the cytoplasm.
Antigenic properties
Viruses are antigenically complex and efficient immunogens, inducing the formation of neutralizing and complement-fixing antigens. The existence of group antigens is likely within species or genera.
Biological properties
Infection
Tailed-viruses are lytic or temperate. Lytic infection results in production of progeny viruses and destruction of the host. Phages adsorb tail-first to specific proteins or polysaccharides on the host cell outer cell surface. In a few cases the primary adsorption sites (receptors) are flagella or pili. Upon adsorption to the outside of the cell, virions undergo complex and often poorly understood rearrangements which release the DNA to enter the cell through the tail. Cell walls are often locally digested by a virion-associated peptidoglycan hydrolase and viral DNA enters the cytoplasm by as yet unknown mechanisms. In some cases DNA entry is stepwise and transcription of the first DNA to enter is required for entry of the rest of the DNA. Empty virions remain outside the infected bacterium, however most viruses inject specific proteins with the DNA. Temperate viruses can, upon infection, either enter a lytic growth cycle (above) or establish a lysogenic state (below). Physiological factors in the cell can affect the decision between these two pathways.
Latency
All three-tailed virus families include genera or species of temperate viruses. Viral genomes in lysogenized cells are called “prophages”. Prophages are either integrated into host cell chromosomes or persist as extrachromosomal elements (plasmids). Integration is usually mediated by recombinases called integrases. The most common are in the tyrosine-active site class and some are in the serine-active site class. For the Mu-like viruses, integration is accomplished by transposases. Integrated prophages typically express only a very small fraction of their genes. The genes that are expressed from the prophage are called “lysogenic conversion” or “cargo” genes, and their products usually alter the properties of the bacterial host. Among these genes is the prophage repressor gene, whose product binds operators in the prophage genome to keep the lytic cascade of gene expression from initiating. Plasmid prophages typically express many of their early genes, some of which are involved in replication of the plasmid (which can be circular or linear). Prophages can often be induced to initiate a lytic growth cycle; DNA damaging agents such as ultraviolet light or mitomycin C cause many prophages to induce.
Host range
Tailed viruses have been found in over 140 prokaryote genera representing most branches of the bacterial and archaeal phylogenetic trees. The host specificity of these viruses can vary widely; some can infect members of multiple genera, but perhaps more common (especially in the host family Enterobacteriaceae, where the most varieties have been studied) are viruses that are specific for particular isolates or groups of isolates of closely related host species.
Transmission in nature
Virions are typically carried and transmitted in aqueous environments, although a few are stable to drying. Virus genomes can be carried as prophages inside host bacteria. Such lysogenic bacteria can induce release of virions, either spontaneously or in response to specific environmental signals.
Geographic distribution
Tailed phages are the most abundant type of organism on Earth; the current best estimates are 1031 particles in our biosphere. If all these phages were laid end to end the line would extend for 2×108 light years. Data from genome sequence analyses imply that these viruses can move around the globe on a time scale that is short relative to the rate at which they accumulate mutations. They have a worldwide distribution and presumably share the habitats of their hosts. An important habitat is inside lysogenic bacteria as prophages.
Phylogenetic relationships within the order and the perils of mosaicism
The recent availability of high-throughput DNA sequencing has led to a dramatic increase in the number of complete genome sequences that are available for members of the Caudovirales. At the latest count 101 myoviruses, 91 podoviruses and 244 siphoviruses are listed in the RefSeq Genomes section of NCBI. The new data substantially enrich our appreciation of the genetic structure and diversity of the global Caudovirales population and of the evolutionary mechanisms within that order. The new data also substantially complicate considerations of how best to represent these viruses in a coherent and easy to use taxonomy.
The hallmark of the genomes of these viruses is that they are genetic mosaics, a property that becomes apparent when two or more genome sequences are compared. The modules of sequence that constitute the mosaic are typically individual genes, but they can also be parts of genes corresponding to protein domains, or small groups of genes such as prohead assembly genes. The mosaicism is evidently the result of non-homologous recombination during the evolution of these viruses. The novel juxtapositions of sequence produced in this way are spread through the population and reassorted with each other by means of homologous recombination. Regardless of mechanism, the overall result is as if each phage had constituted its genome by picking modules from a menu, choosing one module from each of perhaps fifty columns, each of which has alternative choices.
While there is no doubt that recombinational exchange has muddied the relationship between certain phages, recent whole genome comparative proteomics revealed that in many cases clear phylogenetic relationships exist even though DNA sequence similarity is small. Using CoreGenes, a BLASTP-based comparative genomic tool, all fully sequenced members of the Podoviridae and Myoviridae were analyzed. From these studies it was possible to identify high level relationships between phages that shared ≥40% homologous proteins distributed over the length of their genomes (representing phage within the same genus) and lower level relationships in which only 20–30% of the proteins were significantly (BLAST score ≥75) similar. This has led, for example, to the creation of three distinct genera within a new subfamily (Autographivirinae) for phages previously known as the “T7 superfamily” and has helped classify or re-classify many other members of the families Podoviridae and Myoviridae. While this approach has significantly reduced the number of unclassified viruses, there are still many that remain genomic orphans. It has not yet been possible to place certain phages which are clearly related but only at a low level. For example, the myoviruses that infect Prochlorococcus and Synechococcus possess a set of genes which they share with T4-like phages but score very low in CoreGenes. Higher-level relationships will therefore have to be addressed in the future. The tentative classification of phages based purely on morphological grounds and minimal sequence analysis should be discouraged in favor of full sequence analysis on genomes that have been carefully annotated.
In the current ICTV taxonomy, presented here, the division of the order Caudovirales into three families is based solely on tail morphology: members of the family Siphoviridae have long non-contractile tails, Myoviridae have long contractile tails, and Podoviridae have short tails. As might be expected from the discussion above, this hierarchical division of phages on the basis of one character leads to many examples of inappropriate divisions of other characters. One well-known and easily illustrated example of this is shown in Figure 2, comparing phages lambda and P22. These two phages are considered by many phage biologists to be closely related, because they share genome organization (including regulation and layout of transcription and functional order of genes), temperate lifestyle, a number of similarities of gene sequences and they can form viable hybrids. Despite these similarities, they are classified into different families (Siphoviridae and Podoviridae for lambda and P22, respectively) based on their differences in tail morphology. It may be arguable whether the similarities between these two phages are enough for them to be classified in the same family, but it is in any case clear that P22 is much closer to lambda than it is to most other members of the family Podoviridae, such as phages T7 and N4, which have essentially no similarity to lambda in sequence, genome organization, or lifestyle.
Where mosaicism is extensive, ICTV will have to come to terms with the fact that not all phages will be simply classifiable in a straightforward hierarchical manner. Because of this, the ICTV considers the taxonomy of this group to be provisional, and this is the reason that the names of the genera are in a non-official vernacular format. Discussions are ongoing both within the ICTV and in the virology community at large, and there may well be significant changes to the Caudovirales taxonomy in the future, in response to our new understanding of the biology.
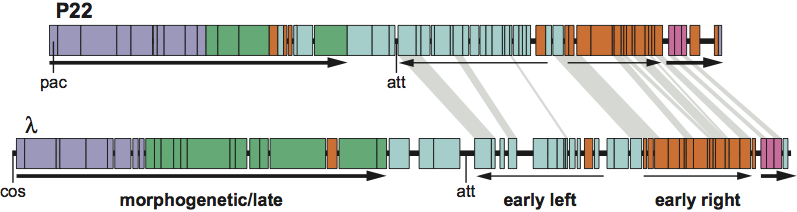 _
_
Figure 2. The mosaic relationship between the genomes of phages P22 and lambda. The circular maps are opened for linear display between the lysis and head genes. The genes in each genome are represented by rectangles. P22 genes that have sequence similarity to lambda genes are connected by light gray trapezoids. The thin arrows represent transcription of the early operons and thick arrows transcription of the late operons. The circular phage genomes are opened at their attachment (att) sites for insertion of the prophage into the host chromosome in lysogens. DNA packaging initiation sites (called pac and cos in P22 and lambda, respectively) are also indicated below the maps.
Similarity with other taxa
Tailed bacterial viruses resemble members of the family Tectiviridae by the presence of a dedicated structure for DNA injection, but differ from them by the permanent nature of their tails and lack of a lipid bilayer. Tailed viruses resemble viruses belonging to the family Herpesviridae in morphogenesis (use of scaffolding proteins, packaging of DNA into preformed shells, maturation of procapsids by proteolytic cleavage, and capsid conformational change) and overall strategy of replication. In addition, temperate tailed phages and members of the family Herpesviridae are able to establish latent infections.
Derivation of names
Caudo: from Latin cauda, “tail”.
Myo: from Greek my, myos, “muscle”, referring to the contractile tail.
Sipho: from Greek siphon, “tube”, referring to the long tail.
Podo: from Greek pous, podos, “foot”, referring to the short tail.
Further reading
Ackermann, H.-W. (2009). Phage classification and characterization. Methods Mol. Biol., 501, 127-140.
Casjens, S. (2005). Comparative genomics and evolution of the tailed-bacteriophages Curr Opin. Microbiol., 8, 451-458.
Hatfull, G.F., Cresawn, S.G. and Hendrix, R.W. (2008). Comparative genomics of the mycobacteriophages: insights into bacteriophage evolution. Res. Microbiol.. 159, 332-339.
Glazko, G., Makarenkov, V. and Mushegian, A. (2007). Evolutionary history of bacteriophages with double-stranded DNA genomes. Biol. Direct., 2, 36.
Lavigne, R., Darius, P., Summer, E.J., Seto, D., Mahadevan, P., Nilsson, A.S., Ackermann, H.W. and Kropinski, A.M. (2009). Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol., 9, 224.
Lavigne, R., Seto, D., Mahadevan, P., Ackermann, H.W. and Kropinski, A.M. (2008). Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol., 159, 406-414.
Lima-Mendez, G., Van Helden, J., Toussaint, A and Leplae, R. (2008). Reticulate representation of evolutionary and functional relationships between phage genomes. Mol. Biol. Evol. 25, 762-777.
Rohwer, F. and Edwards, R. (2002). The Phage Proteomic Tree: a genome-based taxonomy for phage. J. Bacteriol., 184, 4529-4535.
Susskind, M.M. and Botstein, D. (1978). Molecular genetics of bacteriophage P22. Microbiol. Rev., 42, 385-413.
Contributed by
Lavigne, R., Molineux, I.J. and Kropinski, A.M.
The authors acknowledge the contribution to the Eighth ICTV Report of Casjens, S.R. and Hendrix, R.W.
