Family: Baculoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
One or two virion phenotypes are involved in baculovirus infections. Infection is initiated in the gut epithelium by a virus phenotype occluded in a crystalline protein matrix which may be: (a) polyhedral in shape, typically ranging in size from 0.5 to 5 µm and containing many virions for the genera Alphabaculovirus, Gammabaculovirus, Deltabaculovirus (Figure 1 and 2), or (b) ovicylindrical (about 0.3×0.5 µm) and containing only one, or rarely two or more virions (genus Betabaculovirus) (Figure 1). Virions within occlusions consist of one or more rod-shaped nucleocapsids that have a distinct structural polarity and are enclosed within an envelope. For occluded virions, nucleocapsid envelopment occurs within the nucleus (genus Alphabaculovirus) or in the nuclear-cytoplasmic milieu after loss of the nuclear membrane (genus Betabaculovirus). Nucleocapsids average 30–60 nm in diameter and 250–300 nm in length. Spike-like structures have not been reported on envelopes of the occlusion-derived virions (ODV). Virions of the second phenotype (termed budded virions or BV) are generated when nucleocapsids bud through the plasma membrane at the surface of infected cells. BVs typically contain a single nucleocapsid. Their envelopes are derived from the cellular plasma membrane and characteristically appear as a loose-fitting membrane that contains an envelope fusion glycoprotein (EFP), such as the GP64 and F proteins at one end of the virion (see “Proteins”, below) (Figure 1).
Physicochemical and physical properties
ODV buoyant density in CsCl is 1.18–1.25 g cm−3, and that of the nucleocapsid is 1.47 g cm−3. BV buoyant density in sucrose is 1.17–1.18 g cm−3. Virions of both phenotypes are sensitive to organic solvents and detergents. BV is marginally sensitive to heat and pH 8–12, inactivated by pH 3.0, and stable in Mg++ (10−1 M to 10−5 M).
Nucleic acid
Nucleocapsids contain one molecule of circular supercoiled dsDNA, 80–180 kbp in size.
Proteins
Genomic analyses suggest that baculoviruses encode 100–200 proteins. Virions may contain 40 or more different polypeptides. Nucleocapsids from both virion phenotypes (ODV and BV) contain a major capsid protein, a basic DNA binding protein complexed with the viral genome, and at least 2–3 additional proteins. BVs contain an envelope fusion protein (EFP). The EFPs identified to date include the major envelope glycoprotein (the peplomer protein) GP64, which is present in Autographa californica multiple nucleopolyhedrovirus (AcMNPV) and close relatives within the alphabaculoviruses. Most of the alphabaculoviruses and betabaculoviruses encode and appear to utilize F proteins (homologs of the Ld130 protein from Lymantria dispar MNPV (LdMNPV) and the Se8 protein from Spodoptera exigua MNPV (SeMNPV)) as EFP. Several ODV envelope proteins have been identified. Six ODV proteins, including P74, PIF-1, PIF-2, PIF-3, AC96 (PIF-4) and ODV-E56 (PIF-5), are essential for oral infectivity of ODV. The major protein of the occlusion body matrix is a virus-encoded polypeptide of 25–33 kDa. This protein is called polyhedrin for nucleopolyhedroviruses (the former name for the alpha-, delta- and gammabaculoviruses) and granulin for betabaculoviruses. The occlusion body is surrounded by an envelope that contains at least one major protein. The polyhedrin protein of deltabaculoviruses is serologically and genetically unrelated to occlusion body proteins of the alpha-, beta- and gammabaculoviruses.
Lipids
Lipids are present in the envelopes of ODV and BV. Lipid composition differs between the two virion phenotypes.
Carbohydrates
Carbohydrates are present as glycoproteins and glycolipids.
Genome organization and replication
Circular genomic DNA is infectious, suggesting that after cellular entry and uncoating, no virion-associated proteins are not essential for infection. Genomes encode 100–200 proteins (Figure 3). Thirty gene homologs, the so-called baculovirus core genes, are shared among alpha-, beta-, gamma- and deltabaculoviruses. The alphabaculoviruses appear to share 60 homologs, comprising a core group of alphabaculovirus genes. These conserved genes are involved in various functions, including DNA replication, late gene transcription and virion structure. In some cases, larger genome sizes may result from the presence of families of repeated genes. Transcription of baculovirus genes is temporally regulated, and two main classes of genes are recognized: early and late. Late genes may be further subdivided as late and very late. Gene classes (early, late and very late) are not clustered on the baculovirus genome, and both strands of the genome are involved in coding functions. Early genes are transcribed by host RNA polymerase II, while late and very late genes are transcribed by an alpha-amanitin-resistant viral RNA polymerase. RNA splicing occurs, but appears to be rare since only two instances have been identified. Transient early and late gene transcription and DNA replication studies suggest that at least three virus encoded proteins regulate early gene transcription, while approximately 19 viral encoded proteins known as late expression factors (LEFs) are necessary for late gene transcription. Of the approximately 19 LEFs, half appear to be involved in DNA replication. Late gene transcription initiates within or near a highly conserved 5′-TAAG-3′ sequence, which appears to be an essential core element of the baculovirus late promoter. Putative replication origins consist of repeated sequences found at multiple locations within the baculovirus genome. These sequences, termed homologous repeat (hr) regions, do not appear to be highly conserved among different baculovirus species. Single copy, non-hr putative replication origins have also been identified. DNA replication is required for late gene transcription. Most virion structural proteins are encoded by late genes. While transcription of late and very late genes appears to begin immediately after DNA replication, some very late genes that encode occlusion body-specific proteins are transcribed at extremely high levels at a later time. BV production occurs primarily during the late phase, and occlusion body production occurs during the very late phase.
In infected animals, viral replication begins in the insect midgut. Following ingestion, occlusion bodies are solubilized in the gut lumen, releasing the ODVs, which are thought to enter the target epithelial cells via fusion with the plasma membrane at the cell surface. In lepidopteran insects, viral entry into midgut cells occurs in an alkaline environment, up to pH 12. Infection of the midgut is required for initiation of infection in the animal. In most cases, the virus is believed to undergo one round of replication in the midgut epithelium prior to transmission of infection to secondary tissues within the hemocoel. A mechanism for direct movement from the midgut to the hemocoel has also been proposed. DNA replication takes place in the nucleus. In betabaculovirus-infected cells, the integrity of the nuclear membrane is lost during the replication process. With some baculoviruses, replication is restricted to the gut epithelium and progeny virions become enveloped and occluded within these cells, and may be shed into the gut lumen with sloughed epithelium, or released upon death of the host. In other baculoviruses, the infection is transmitted to internal organs and tissues. The second virion phenotype, BV, which buds from the basolateral membrane of infected gut cells is required for transmission of the infection into the hemocoel. In secondarily infected tissues, BV is produced during the late phase and ODV is produced during the very late phase of the infection. Infected fat body cells are the primary location of occluded virus production in lepidopteran insects. Occluded virus matures within nuclei of infected cells for alpha-, gamma- and deltabaculoviruses and within the nuclear-cytoplasmic milieu for betabaculoviruses. Occlusion bodies containing infectious ODV virions are released upon death, and usually liquefaction, of the host.
Antigenic properties
Antigenic determinants that cross-react between different baculoviruses exist on virion proteins and on the major occlusion body polypeptide: polyhedrin or granulin. Neutralizing antibodies react with the major surface glycoprotein of BV.
Biological properties
Baculoviruses have been isolated from insects only; primarily from insects of the order Lepidoptera, but also Hymenoptera, and Diptera. Transmission: naturally (i) horizontal transmission by contamination of food, egg surface, etc. with occlusion bodies; (ii) vertical transmission within the egg either from infected female or male adults; experimentally (iii) by injection of intact hosts with BV; (iv) by infection or transfection of cell cultures. Typically the infection process in a permissive insect host requires approximately one week, and as an end result, the diseased insect liquefies releasing infectious occlusion bodies into the environment. Occlusion bodies represent an environmentally stable form of the virus with increased resistance to chemical and physical decay as well as inactivation by UV light.
Genus Alphabaculovirus
Type species Autographa californica multiple nucleopolyhedrovirus
Distinguishing features
Virions of the ODV phenotype are embedded within an occlusion body composed of a crystalline matrix of a single viral protein (polyhedrin). Each occlusion body measures 0.15 to 15 µm in size, matures within nuclei of infected cells and characteristically contains many enveloped virions. The occluded virions are packaged with either single (S) or multiple (M) nucleocapsids within a single viral envelope. Some virus species manifest both phenotypes. Factors that regulate nucleocapsid packaging are unknown and, for some species, packaging may be variable. S and M designations in common usage have been retained for species where variability has not been reported and for distinct viruses that would otherwise have identical designations under the current nomenclature. Nucleocapsids are rod-shaped (30–60 nm×250–300 nm) and contain a single molecule of circular supercoiled dsDNA of 110–170 kbp in size. Nucleocapsid length appears to be proportional to genome size. During viral entry, nucleocapsids are believed to be transported through the nuclear membrane and into the nucleus, where uncoating and viral replication occur. Hosts include one order of insects, the Lepidoptera.
Species demarcation criteria in the genus
Because detailed comparative data are lacking in most cases, species parameters are not well defined. However, species distinctions indicated here are broadly based on host range and specificity, DNA restriction profiles, DNA sequences from various regions of the genome, and predicted protein sequence similarities.
List of species in the genus Alphabaculovirus
| Adoxophyes honmai nucleopolyhedrovirus |
|
|
| Adoxophyes honmai nucleopolyhedrovirus (ADN001) | [AP006270=NC_004690] | (AdhoNPV) |
| Agrotis ipsilon multiple nucleopolyhedrovirus |
|
|
| Agrotis ipsilon nucleopolyhedrovirus (Illinois) | [EU839994=NC_011345] | (AgipNPV) |
| Anticarsia gemmatalis multiple nucleopolyhedrovirus |
|
|
| Anticarsia gemmatalis multiple nucleopolyhedrovirus (2D) | [DQ813662=NC_008520] | (AgMNPV) |
| Autographa californica multiple nucleopolyhedrovirus |
|
|
| Autographa californica multiple nucleopolyhedrovirus (C6) | [L22858=NC_001623] | (AcMNPV) |
| Galleria mellonella multiple nucleopolyhedrovirus |
| (GmMNPV) |
| Plutella xylostella multiple nucleopolyhedrovirus (CL3) | [DQ457003=NC_008349] |
|
| Spodoptera exempta multiple nucleopolyhedrovirus |
| (SpexNPV) |
| Trichoplusia ni multiple nucleopolyhedrovirus |
| (TnMNPV) |
| Bombyx mori nucleopolyhedrovirus |
|
|
| Bombyx mori nucleopolyhedrovirus (T3) | [L33180=NC_001962] | (BmNPV) |
| Bombyx mandarina nucleopolyhedrovirus (S1) | [FJ882854=NC_012672] | (BomaNPV) |
| Buzura suppressaria nucleopolyhedrovirus |
|
|
| Buzura suppressaria nucleopolyhedrovirus (S13) | [U61154.1=GI_2138112] | (BuzuNPV) |
| Choristoneura fumiferana DEF multiple nucleopolyhedrovirus |
|
|
| Choristoneura fumiferana DEF multiple nucleopolyhedrovirus | [AY327402=NC_005137] | (CfDefNPV) |
| Choristoneura fumiferana multiple nucleopolyhedrovirus |
|
|
| Choristoneura fumiferana multiple nucleopolyhedrovirus (Ireland) | [AF512031=NC_004778] | (CfMNPV) |
| Choristoneura rosaceana nucleopolyhedrovirus |
|
|
| Choristoneura rosaceana nucleopolyhedrovirus |
| (ChroNPV) |
| Ecotropis obliqua nucleopolyhedrovirus |
|
|
| Ecotropis obliqua nucleopolyhedrovirus (A1) | [DQ837165=NC_008586] | (EcobNPV) |
| Epiphyas postvittana nucleopolyhedrovirus |
|
|
| Epiphyas postvittana nucleopolyhedrovirus | [AY043265=NC_003083] | (EppoNPV) |
| Heliocoverpa armigera nucleopolyhedrovirus |
|
|
| Heliocoverpa armigera nucleopolyhedrovirus (C1) | [AF303045=NC_003094] | (HearNPV-C1) |
| Heliocoverpa armigera nucleopolyhedrovirus (NNG1) | [AP010907=NC_011354] | (HearNPV-NNG1) |
| Heliocoverpa armigera nucleopolyhedrovirus (G4) | [AF271059=NC_002654] | (HearNPV-G4) |
| Helicoverpa zea single nucleopolyhedrovirus |
|
|
| Helicoverpa zea single nucleopolyhedrovirus | [AF334030=NC_003349] | (HzSNPV) |
| Lymantria dispar multiple nucleopolyhedrovirus |
|
|
| Lymantria dispar multiple nucleopolyhedrovirus | [AF081810=NC_001973] | (LdMNPV) |
| Mamestra brassicae multiple nucleopolyhedrovirus |
|
|
| Mamestra brassicae multiple nucleopolyhedrovirus (Oxford) |
| (MbMNPV) |
| Mamestra configurata nucleopolyhedrovirus A |
|
|
| Mamestra configurata nucleopolyhedrovirus A (90/2) | [U59461=NC_003529] | (MacoNPV-A) |
| Mamestra configurata nucleopolyhedrovirus A (90/4) | [AF539999] |
|
| Mamestra configurata nucleopolyhedrovirus B |
|
|
| Mamestra configurata nucleopolyhedrovirus B (96B) | [AY126275=NC_004117] | (MacoNPV-B) |
| Orgyia pseudotsugata multiple nucleopolyhedrovirus |
|
|
| Orgyia pseudotsugata multiple nucleopolyhedrovirus | [U75930=NC_001875] | (OpMNPV) |
| Spodoptera exigua multiple nucleopolyhedrovirus |
|
|
| Spodoptera exigua multiple nucleopolyhedrovirus (US) | [AF169823=NC_002169] | (SeMNPV) |
| Spodoptera frugiperda multiple nucleopolyhedrovirus |
|
|
| Spodoptera frugiperda multiple nucleopolyhedrovirus (3AP2) | [EF035042=NC_009011] | (SfMNPV) |
| Spodoptera littoralis nucleopolyhedrovirus |
|
|
| Spodoptera littoralis nucleopolyhedrovirus (M2) |
| (SpliNPV) |
| Spodoptera litura nucleopolyhedrovirus |
|
|
| Spodoptera litura nucleopolyhedrovirus G2 | [AF325155=NC_003102] | (SpltNPV) |
| Thysanoplusia orichalcea nucleopolyhedrovirus |
|
|
| Thysanoplusia orichalcea nucleopolyhedrovirus A28 |
| (ThorNPV) |
| Trichoplusia ni single nucleopolyhedrovirus |
|
|
| Trichoplusia ni single nucleopolyhedrovirus | [DQ017380=NC_007383] | (TnSNPV) |
| Wiseana signata nucleopolyhedrovirus |
|
|
| Wiseana signata nucleopolyhedrovirus |
| (WisiNPV) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses, which may be members of the genus Alphabaculovirus but have not been approved as species
| Adoxophyes orana nucleopolyhedrovirus | [EU591746=NC_011423] | (AdorNPV) |
| Agrotis segetum nucleopolyhedrovirus A | [DQ123841=NC_007921] | (AgseNPV) |
| Anagrapha falcifera multiple nucleopolyhedrovirus |
| (AnfaNPV) |
| Antheraea pernyi nucleopolyhedrovirus | [DQ486030=NC_008035] | (AnpeNPV) |
| Chrysodeixis chalcites nucleopolyhedrovirus | [AY864330=NC_007151] | (ChchNPV) |
| Clanis bilineata nucleopolyhedrovirus | [DQ504428=NC_008293] | (ClbiNPV) |
| Euproctis pseudoconspersa nucleopolyhedrovirus | [DQ837165=NC_008586] | (EupsNPV) |
| Hyphantria cunea nucleopolyhedrovirus | [AP009046=NC_007767] | (HycuNPV) |
| Leucania separata nucleopolyhedrovirus | [AY394490=NC_008348] | (LeseNPV) |
| Maruca vitrata nucleopolyhedrovirus | [EF125867=NC_008725] | (MaviNPV) |
| Orgyia leucostigma nucleopolyhedrovirus | [EU309041=NC_010276] | (OrleNPV) |
| Orgyia pseudotsugata single nucleopolyhedrovirus |
| (OpSNPV) |
| Panolis flammea nucleopolyhedrovirus |
| (PaflNPV) |
| Rachiplusia ou multiple nucleopolyhedrovirus | [AY145471=NC_004323] | (RoMNPV) |
Genus Betabaculovirus
Type species Cydia pomonella granulovirus
Distinguishing features
Two virion phenotypes (BV and ODV) may be characteristic of a virus species. One (ODV) is occluded within an ovicylindrical occlusion body composed mainly of a single protein (granulin), which is a homolog (ortholog) to polyhedrin of alpha- and gammabaculoviruses. Each occlusion body measures approximately 0.13×0.50 µm in size and characteristically contains one virion. Each ODV virion typically contains a single nucleocapsid within a single envelope. Occluded virions may mature among nuclear-cytoplasmic cellular contents after loss of the nuclear membrane of infected cells. Nucleocapsids are rod-shaped (30–60 nm×250–300 nm) and contain a single molecule of circular supercoiled dsDNA, approximately 110–180 kbp in size. Uncoating is thought to occur by a mechanism in which viral DNA is extruded into the nucleus through the nuclear pore while the capsid remains in the cytoplasm. Species of this genus have been isolated only from the insect order Lepidoptera.
Species demarcation criteria in the genus
Refer to genus Alphabaculovirus.
List of species in the genus Betabaculovirus
| Adoxophyes orana granulovirus |
|
|
| Adoxophyes orana granulovirus | [AF547984=NC_005038] | (AdorGV) |
| Artogeia rapae granulovirus |
|
|
| Artogeia rapae granulovirus |
| (ArGV) |
| Pieris brassicae granulovirus (384) |
| (PbGV) |
| Choristoneura fumiferana granulovirus |
|
|
| Choristoneura fumiferana granulovirus (Bonaventure) |
| (ChfuGV) |
| Cryptophlebia leucotreta granulovirus |
|
|
| Cryptophlebia leucotreta granulovirus (CV3) | [AY229987=NC_005068] | (CrleGV) |
| Cydia pomonella granulovirus |
|
|
| Cydia pomonella granulovirus (M1) | [U53466=NC_002816] | (CpGV) |
| Harrisina brillians granulovirus |
|
|
| Harrisina brillians granulovirus (M2) |
| (HabrGV) |
| Helicoverpa armigera granulovirus |
|
|
| Helicoverpa armigera granulovirus | [EU255577=NC_010240] | (HearGV) |
| Lacanobia oleracea granulovirus |
|
|
| Lacanobia oleracea granulovirus (S1) |
| (LaolGV) |
| Phthorimaea operculella granulovirus |
|
|
| Phthorimaea operculella granulovirus | [AF499596=NC_004062] | (PhopGV) |
| Plodia interpunctella granulovirus |
|
|
| Plodia interpunctella granulovirus (B3) |
| (PiGV) |
| Plutella xylostella granulovirus |
|
|
| Plutella xylostella granulovirus (K1) | [AF270937=NC_002593] | (PlxyGV) |
| Pseudalatia unipuncta granulovirus |
|
|
| Pseudalatia unipuncta granulovirus (Hawaiian) | [EU678671=NC_013772] | (PsunGV) |
| Trichoplusia ni granulovirus |
|
|
| Trichoplusia ni granulovirus (M10-5) |
| (TnGV) |
| Xestia c-nigrum granulovirus |
|
|
| Xestia c-nigrum granulovirus (alpha4) | [AF162221=NC_002331] | (XecnGV) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses, which may be members of the genus Betabaculovirus but have not been approved as species
| Agrotis segetum granulovirus (Xinjiang) | [AY522332=NC_005839] | (AgseGV) |
| Choristoneura occidentalis granulovirus | [DQ333351=NC_008168] | (ChocGV) |
| Spodoptera litura granulovirus (K1) | [DQ288858=NC_009503] | (SpliGV) |
Genus Gammabaculovirus
Type species Neodiprion lecontei nucleopolyhedrovirus
Distinguishing features
The virions are occluded singly into the viral occlusion bodies. The virus is restricted to the host midgut and causes what was previously described in the literature as “infectious diarrhea”. Genome sequencing analyses from three viruses (NeleNPV, NeseNPV, NeabNPV) revealed that these viruses do not encode typical envelope fusion proteins found in other baculoviruses. This raised the question of whether the budded virus phenotype plays a role in Gammabaculovirus biology. Also, in comparison to other baculoviruses, the genomes of members of the genus Gammabaculovirus are relatively high in A+T content (on the order of 67%). The genomes of the three sequenced gammabaculoviruses are collinear except for a large non-syntenic region between the DNA polymerase and polyhedrin genes. This region contains genes and ORFs not shared among the three characterized genomes.
Species demarcation criteria in the genus
Refer to genus Alphabaculovirus.
List of species in the genus Gammabaculovirus
| Neodiprion lecontei nucleopolyhedrovirus |
|
|
| Neodiprion lecontei nucleopolyhedrovirus | [AY349019=NC_005906] | (NeleNPV) |
| Neodiprion sertifer nucleopolyhedrovirus |
|
|
| Neodiprion sertifer nucleopolyhedrovirus | [AY430810=NC_005905] | (NeseNPV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses, which may be members of the genus Gammabaculovirus but have not been approved as species
| Gilpinia hercyniae nucleopolyhedrovirus (i7) |
| (GiheNPV) |
| Neodiprion abietis nucleopolyhedrovirus | [DQ317692=NC_008252] | (NeabNPV) |
Genus Deltabaculovirus
Type species Culex nigripalpus nucleopolyhedrovirus
Distinguishing features
Replication of Culex nigripalpus nucleopolyhedrovirus (CuniNPV) is restricted to host midgut epithelium, primarily in larval stages but rarely in adults. Two virion phenotypes may be characteristic of a virus species. Virions of the ODV phenotype are embedded within an occlusion body composed of a crystalline matrix of a single viral protein with no homology to polyhedrin or granulin of other baculovirus genera. Each occlusion body ranges in size from 0.5 to 15 µm and contains few (1–4) or many (50+) singly enveloped virions depending on the strain of virus, lacks the polyhedron envelope of other baculoviruses and matures within nuclei of infected cells. Nucleocapsids are rod-shaped (30–60 nm×200–250 nm) and contain a single molecule of circular supercoiled dsDNA. Transmission of CuniNPV to larval mosquitoes is strongly influenced by divalent cations: Mg2+ is a potent enhancer of transmission while Ca2+ is a strong inhibitor. The CuniNPV genome is 108,252 bp and encodes at least 109 putative proteins, some of which have sequence homology with those from other baculoviruses. Homologous proteins are involved in early and late gene expression, DNA replication, as well as structural and auxiliary functions. Gene orientation and order in the genome of CuniNPV is different from other baculovirus genera. The CuniNPV genome lacks genes with homologs for several essential and stimulatory genes for DNA replication and transcription found in other baculovirus genera and also lacks homologs for other conserved structural genes involved in the formation of nucleocapsids and occlusion bodies. Only 36 of the 109 putative CuniNPV predicted proteins demonstrate clear homology to proteins from other baculoviruses, and 72 of the CuniNPV ORFs show no homology to any other known baculovirus ORFs. Hosts include at least three genera of mosquitoes but other mosquito genera and families of Diptera are likely hosts.
Species demarcation criteria in the genus
Refer to genus Alphabaculovirus.
List of species in the genus Deltabaculovirus
| Culex nigripalpus nucleopolyhedrovirus |
|
|
| Culex nigripalpus nucleopolyhedrovirus (Florida 1997) | [AF403738=NC_003084] | (CuniNPV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses, which may be members of the genus Deltabaculovirus but have not been approved as species
| Aedes sollicitans nucleopolyhedrovirus | (AesoNPV) |
| Uranotaenia sapphrinia nucleopolyhedrovirus | (UrsaNPV) |
List of unassigned species in the family
None reported.
Phylogenetic relationships within the family
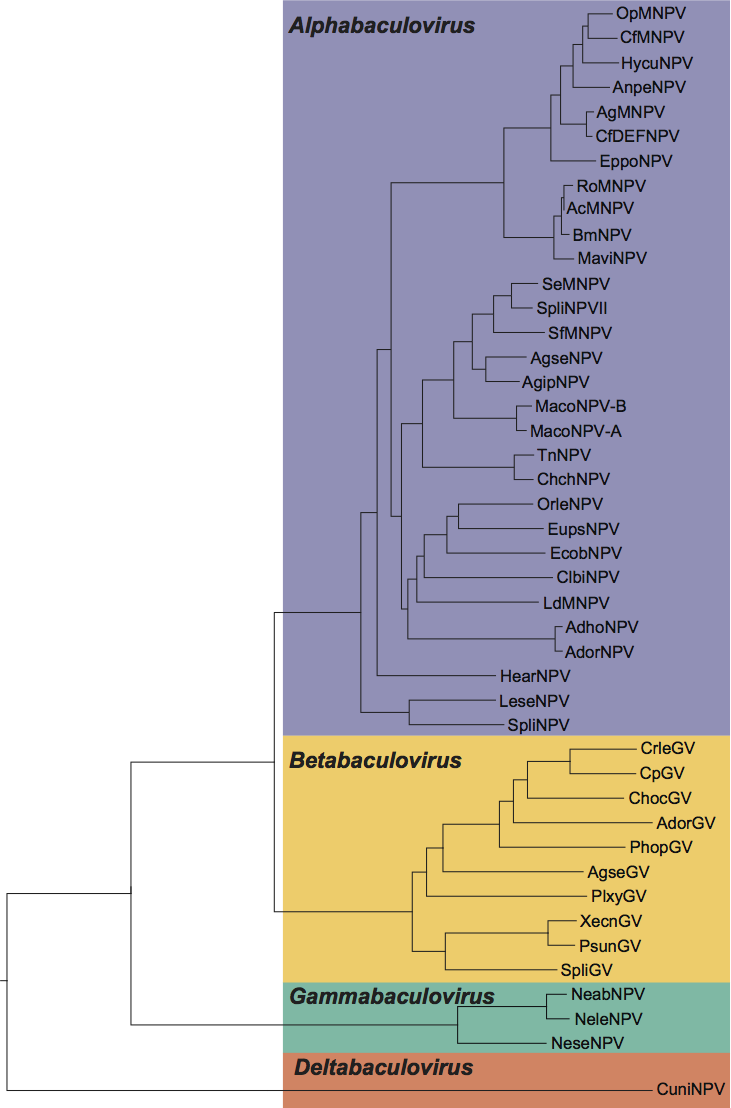
Phylogenetic analysis based on the 30 baculovirus core genes shows that the family comprises four monophyletic groups (Figure 3), which can also be discriminated based on the insect orders of their hosts and on their morphology. Thus the new family structure classifies baculoviruses into four genera Alphabaculovirus, Betabaculovirus, Gammabaculovirus, Deltabaculovirus.
Phylogeny of the Baculoviridae. The maximum likelihood tree, based on the alignment of 30 genes, shows the relationships of the 39 species for which a completely annotated genome was available at time of analyses. Abbreviations are defined in lists of species and related viruses above.
Similarity with other taxa
Members of the family Baculoviridae share structural and biological characters with the unassigned Nudiviruses, which formerly were called “non-occluded” baculoviruses. The nudiviruses share at least 20 core genes with baculoviruses. Baculoviruses also share at least 10 core genes with members of the genus Bracovirus, family Polydnaviridae.
Derivation of names
Baculo: from baculum, meaning “rod”, referring to the morphology of the nucleocapsid.
Granulo: from “granule”, referring to the relatively small size and granular appearance of GV occlusion bodies in infected cells.
Polyhedro: from “polyhedron”, referring to the multifaceted shape of occlusion bodies.
Further reading
Blissard, G.W. (1996). Baculovirus–insect cell interactions. Cytotech., 20, 73-93.
Braunagel, S.C. and Summers, M.D. (2007). Molecular biology of the baculovirus occlusion-derived virus envelope. Curr. Drug Targets, 8, 1084-1095.
Herniou, E.A. and Jehle, J.A. (2007). Baculovirus phylogeny and evolution. Curr. Drug Targets, 8, 1043-1050.
Herniou, E.A., Olszewski, J.A., Cory, J.S. and O'Reilly, D.R. (2003). The genome sequence and evolution of baculoviruses. Ann. Rev. Entomol., 48, 211-234.
Jehle, J.A., Blissard, G.W., Bonning, B.C., Cory, J.S., Herniou, E.A., Rohrmann, G.F., Theilmann, D.A., Thiem, S.M. and Vlak, J.M. (2006). On the classification and nomenclature of baculoviruses: A proposal for revison. Arch. Virol., 151, 1257-1266.
Jehle, J.A., Lange, M., Wang, H., Hu, Z.-H., Wang, Y. and Hauschild, R. (2006). Molecular identification and phylogentic analysis of baculoviruses of Lepidoptera. Virology, 346, 180-196.
Miller, L.K. (Ed.) (1997). The Baculoviruses. Plenum Press, New York.
Moser, B.A., Becnel, J.J., White, S.E., Afonso, C., Kutish, G., Shanker S. and Almira, E. (2001). Morphological and molecular evidence that Culex nigripalpus baculovirus is an unusual member of the family Baculoviridae. J. Gen. Virol., 82, 283-297.
Rohrmann, G.F. (2008). Baculovirus Molecular Biology. Bethesda, MD: National Library of Medicine, NCBI.
van Oers, M.M. and Vlak, J.M. (2007). Baculovirus genomics. Curr. Drug Targets, 8, 1051-1068.
Figures
Figure 1 Baculovirus occlusion bodies, virions and nucleocapsids. (Upper left) The structures of occlusion bodies from baculoviruses in the genera Alphabaculovirus (nucleopolyhedrovirus, NPV) and Betabaculovirus (granulovirus, GV) are illustrated. Virions embedded in nucleopolyhedrovirus occlusion bodies may contain multiple nucleocapsids (MNPV) or single nucleocapsids (SNPV). (Upper right) The two baculovirus virion phenotypes are illustrated as diagrams with shared and phenotype-specific components (from Blissard, 1996). (Bottom) Transmission electron micrographs of occlusion bodies (MNPV, SNPV and GV), virion phenotypes BV (budded virions), ODV (occlusion-derived virions) and nucleocapsids (NC). Nucleopolyhedrovirus occlusion bodies of the MNPV (Autographa californica MNPV, top left) and SNPV (Trichoplusia ni SNPV, top middle) types are compared to granulovirus occlusion bodies (Estigmine acrea GV, top right). Transmission electron micrographs of virions of the BV (Lymantria dispar MNPV, bottom left) and ODV (Autographa californica MNPV, bottom center) phenotypes are shown beside negatively stained nucleocapsids (Autographa californica MNPV, bottom right).
(Electron micrographs courtesy of J.R. Adams [LdMNPV BV virion] and R. Granados [all others].)
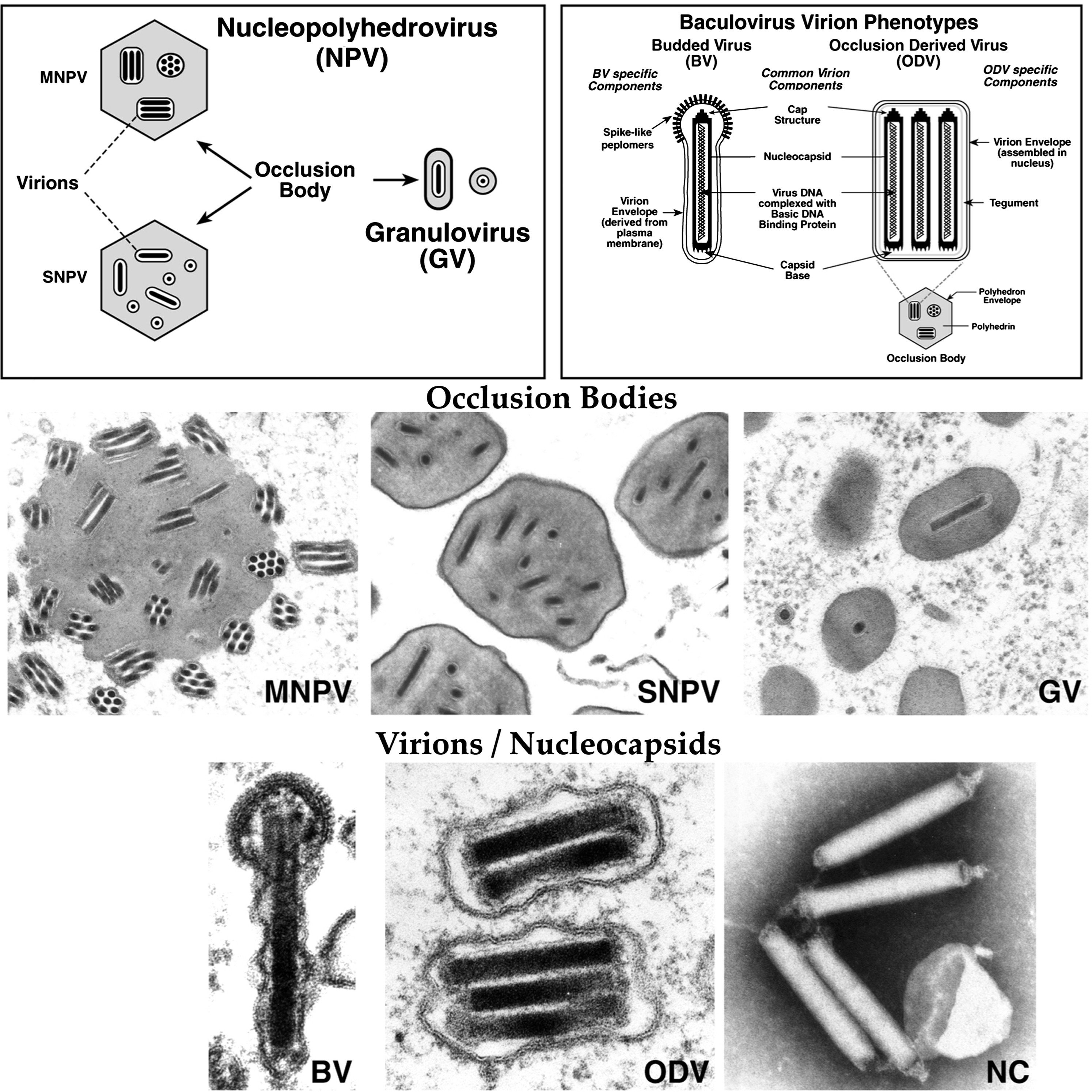
Figure 2 Transmission electron micrographs of occlusion bodies from the genera (A) Gammabaculovirus (Neodiprion abietis nucleopolyhedrovirus, courtesy of C.J. Lucarotti) and (B) Deltabaculovirus
(Culex nigripalpus nucleopolyhedrovirus, courtesy of J.J. Becnel).
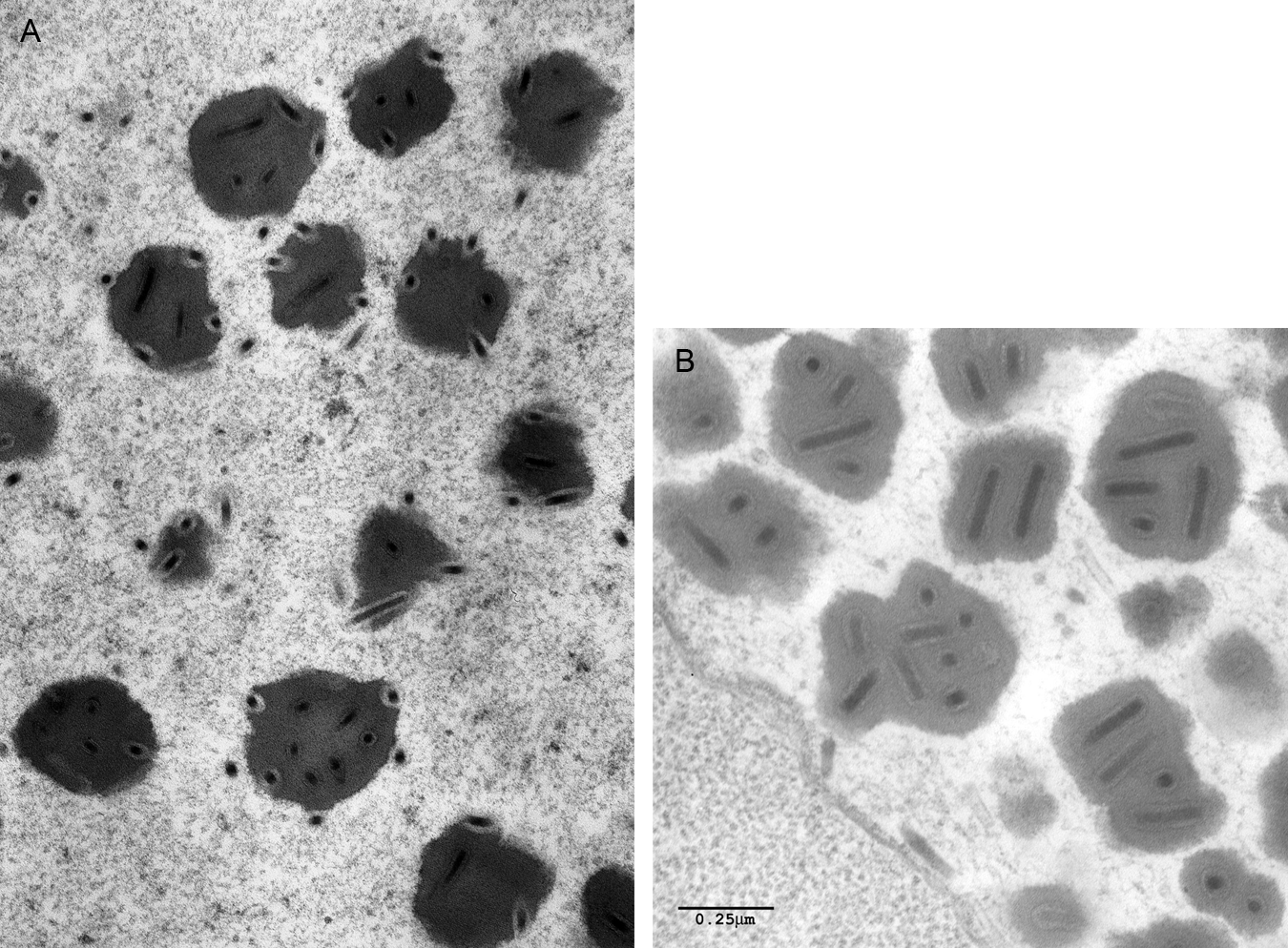
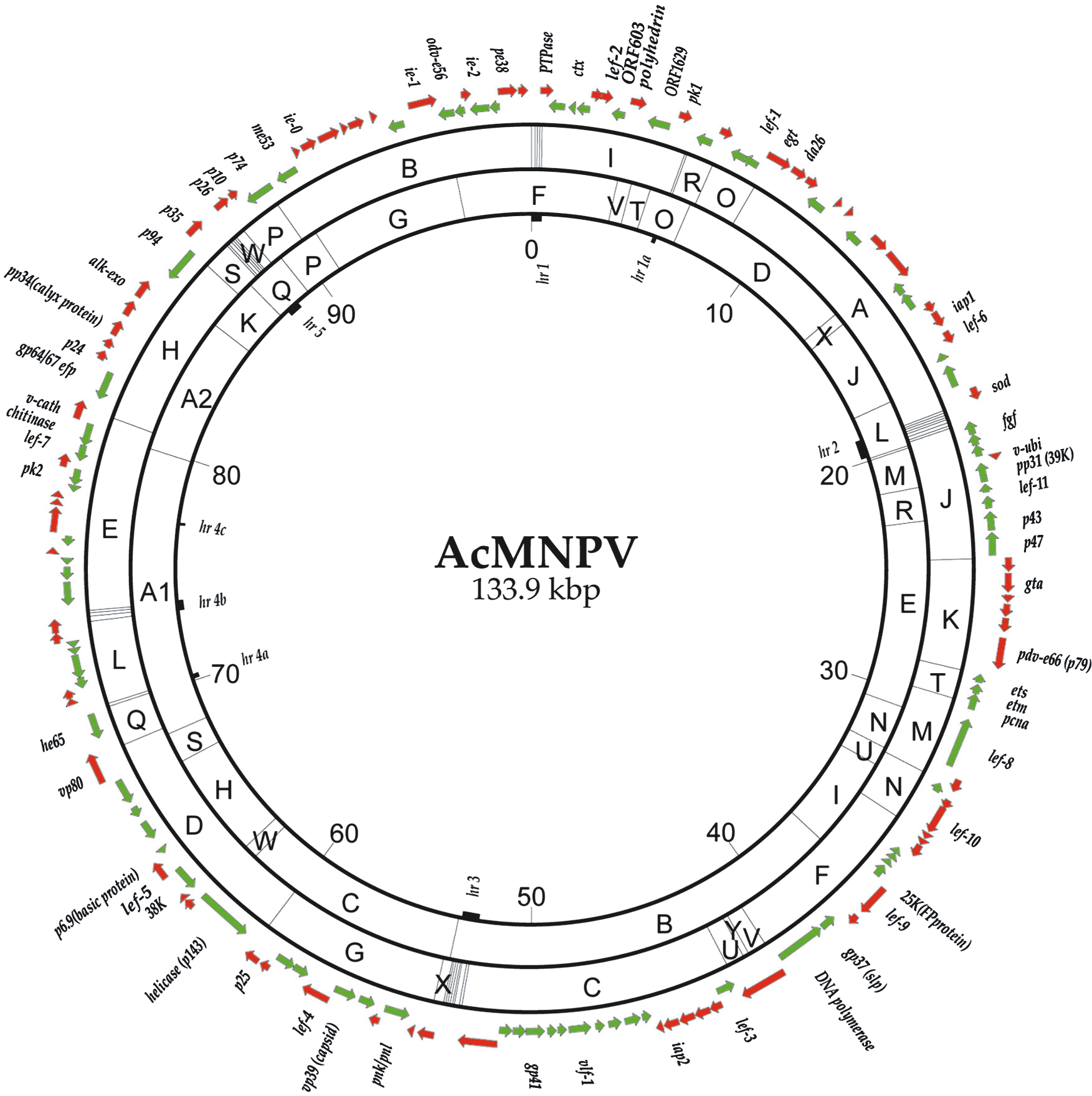
Figure 3 The covalently closed circular genome of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is illustrated with locations and orientations of known and predicted ORFs (arrows). Restriction maps for EcoRI and HindIII are indicated by letters on outer and inner rings, respectively. Locations of homologous repeat (hr) sequences are indicated on the inside of the circle as small filled boxes. Map units are indicated on the inside of the map (1 map unit = 1.339 kbp)
(redrawn from Ayres et al. (1994). Virology, 202, 586605). (See Rohrmann, 2011 for details.)
Figure 4 Phylogeny of the Baculoviridae. The maximum likelihood tree, based on the alignment of 30 genes, shows the relationships of the 39 species for which a completely annotated genome was available at time of analyses. Abbreviations are defined in lists of species and related viruses above.