Updated ICTV Online (10th) Report Chapter Available
Follow this link to the current Ascoviridae Online Report
Family: Ascoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Since only one genus is currently recognized, the family description corresponds to the genus description.
Genus Ascovirus
Type species Spodoptera frugiperda ascovirus 1a
Virion properties
Morphology
Virions of ascoviruses are either bacilliform, ovoidal or allantoid in shape, depending on the species. Ascovirus virions have complex symmetry, and are large, measuring approximately 130 nm in diameter by 200–400 nm in length. The virion consists of an inner particle surrounded by an outer envelope. The inner particle typically measures 80×300 nm and contains a DNA/protein core bounded by an apparent lipid bilayer, the external surface of which bears a distinctive layer of protein subunits. The virion, therefore, appears to contain two lipid membranes, one associated with the inner particle and the other forming the envelope. In negatively stained preparations, virions have a distinctive reticulate appearance thought to result from superimposition of protein subunits on the surface of the internal particle with those in the external envelope (Figure 1).
Physicochemical and physical properties
Virions are sensitive to organic solvents and detergents. Other properties are not known.
Nucleic acid
The inner particle contains a single molecule of circular dsDNA ranging in size from 150 to 190 kbp. G+C content ranges from 42% to 60% depending on the species.
Proteins
Virions contain at least 21 polypeptides ranging in size from 6 to 200 kDa. Ascovirus genomes contain from 117 to 180 genes, of which 40 are common with each other.
Lipids
Ultrastructural evidence and detergent sensitivity indicate the presence of lipid in both the outer envelope and inner particle of the virion. Specific lipid composition is not known.
Carbohydrates
None reported.
Genome organization and replication
The genomes of ascoviruses are composed of circular dsDNA. The genomes of three ascoviruses, Heliothis virescens ascovirus 3e (HvAV-3e), Spodoptera frugiperda ascovirus 1a (SfAV-1a) and Trichoplusia ni ascovirus 6a (TnAV-6a; previously named TnAV2c) have been sequenced. The recent sequencing of the Diadromus pulchellus ascovirus 4a (DpAV-4a) genome has revealed that this virus is not an ascovirus, but originated from a sibling family.
Ascoviruses initiate replication in the nucleus. The nucleus enlarges and ruptures, after which the plasmalemma invaginates, forming internal membranous folds that cleave the cell into a cluster of virion-containing vesicles. Virion assembly becomes apparent after the nucleus ruptures. The first recognizable structural component of the virion to form is the multilaminar layer of the inner particle. Based on its ultrastructure, this layer consists of a unit membrane and an exterior layer of protein subunits. As the multilaminar layer forms, the dense DNA/protein core assembles along the inner surface. As this process continues, the allantoid, ovoidal or bacilliform shape of the inner particle becomes apparent. After the inner particle is assembled, it is enveloped by a membrane, synthesized de novo, or elaborated from cell membranes, within the cell or vesicle. In SfAV-1a, the virions are occluded in an occlusion body composed of minivesicles and protein.
Antigenic properties
No information available.
Biological properties
Host range
Ascoviruses cause disease in lepidopteran larvae and pupae, and have been reported most commonly from species of the family Noctuidae, including Trichoplusia ni, Heliothis virescens, Helicoverpa zea, Spodoptera frugiperda and Autographa precationis. TnAV-2a and HvAV-3a have been shown to have a broad experimental host range among larvae of the lepidopteran family Noctuidae, but the host range of SfAV-1a is restricted primarily to species of Spodoptera. However, ascoviruses could have an expanded host range that includes non-noctuid insects. For example, recent studies have demonstrated that HvAV-3e is able to productively propagate in Crocidomia pavonana and Plutella xylostella larva, lepidopteran species belonging to families, respectively, Crambidae and Plutellidae.
Transmission
Ascoviruses are difficult to transmit per os, and experimental studies as well as field observations indicate most are transmitted horizontally by endoparasitic wasps (Hymenoptera), many species of which belong to the families Braconidae and Ichneumonidae. During egg-laying, the ovipositor of female wasps becomes contaminated with virions and virion-containing vesicles that circulate in the blood (hemolymph) of infected caterpillars. Wasps contaminated in this manner subsequently transmit ascovirus virions to new caterpillar hosts during oviposition.
Geographical distribution
Ascoviruses are known from the United States, Europe, Australia, Indonesia and Mexico, and likely occur worldwide, that is, wherever species of Lepidoptera and their hymenopteran parasites occur. Ascoviruses cause a chronic, fatal disease that markedly retards larval development, but which typically exhibits little other gross pathology. This lack of easily recognizable gross pathology probably accounts for the lack of host records from many geographical regions.
Cytopathic effects
Ascoviruses vary in tissue tropism, with some attacking most host tissues, such as TnAV-2a and HvAV-3a, whereas others, such as the type species SfAV-1a, are restricted to the fat body. The unique property of ascovirus infection is a novel cytopathology in which host cells cleave to form virion-containing vesicles by a developmental process utilizing a modified form of apoptosis. Infection results in nuclear hypertrophy followed by lysis. The anucleate cell enlarges 5–10-fold and then cleaves into 10–30 virion-containing vesicles. Membranes delimiting vesicles form by invagination of the plasmalemma and membrane synthesis. Millions of vesicles (ca. 107–108 vesicles ml−1) accumulate in the hemolymph, turning it milky white. Opaque white hemolymph containing refractile virion vesicles is unique and diagnostic for ascovirus disease.
Species demarcation criteria in the genus
The following list of characters is used in combination to differentiate species in the genus:
- Virion morphology
- Phylogenetic position of genes encoding the major capsid protein and the DNA polymerase and their homologs in other characterized ascoviruses and invertebrate iridoviruses
- Presence or absence of occlusion bodies
- Lack of DNA/DNA hybridization with other species at low stringency
- Restriction enzyme fragment length polymorphisms (RFLPs)
- Host of isolation and experimental host range
- Tissue tropism
- Association with specific hymenopteran parasites, if apparent
Ascoviruses can have broad host ranges among the larvae of lepidopteran species, and the fat body tissue is a major site of replication for most species. In addition, the virions of most isolates are similar in size and shape. The above characters are therefore used in combination to distinguish existing and new ascovirus species from one another. Hybridization studies have proven particularly useful, and when combined with RFLPs and phylogenetics can also be used to distinguish variants within a species.
For example, SfAV-1a DNA does not hybridize with HvAV-3a or TnAV-2a DNAs under conditions of low stringency. TnAV-2a DNA hybridizes to some extent with HvAV-3a DNA, but not as strongly as it does with homologous DNA. In addition, TnAV-2a replicates in a range of larval tissues including the fat body, tracheal matrix and epidermis, but SfAV-1a and HvAV-3a appear to replicate, respectively, only or primarily in the fat body tissue of most hosts. SfAV-1a virions are bacilliform and are occluded in vesiculate occlusion bodies, whereas TnAV-2a virions are allantoid and are not occluded in occlusion bodies. HvAV-3a virions vary from allantoid to bacilliform, and are not occluded in occlusion bodies.
When the genome of a new isolate cross-hybridizes with that of an existing species member, RFLPs can be used to distinguish variants. Numerous ascovirus isolates, for example, have been obtained from larvae of different noctuid species, including Heliothis virescens, Helicoverpa zea, Autographa precationis and Spodoptera exigua in the United States, as well as from Helicoverpa and Spodoptera species in Australia and Indonesia. The DNA of many of these isolates shows strong reciprocal hybridization with HvAV-3a DNA under conditions of high stringency. RFLP profiles of these isolates, however, often show variations from HvAv-3a that range from minor to major. Because these isolates cross-hybridize strongly with HvAV-3a, they are considered variants of this viral species. Moreover, experimentally these isolates have been shown to have host ranges that overlap with HvAV-3a, providing additional evidence that they are variants of the same species. A similar situation occurs with isolates of TnAV-2a and TnAV-2b.
List of species in the genus Ascovirus
| Heliothis virescens ascovirus 3a |
|
|
| Heliothis virescens ascovirus 3a | [AJ279817*] | (HvAV-3a) |
| Heliothis virescens ascovirus 3b |
| (HvAV-3b) |
| Heliothis virescens ascovirus 3c | [AJ312696*, AJ312697*, AJ312704*, AJ312698*] | (HvAV-3c) |
| Heliothis virescens ascovirus 3d | [AJ279820*] | (HvAV-3d) |
| Heliothis virescens ascovirus 3e | [EF133465=NC_009233] | (HvAV-3e) |
| Heliothis virescens ascovirus 3f | [DQ015956*] | (HvAV-3f) |
| Heliothis virescens ascovirus 3g | [AJ620611*, AJ620613*] | (HvAV-3g) |
| (Spodoptera exigua ascovirus 5a) |
|
|
| Spodoptera frugiperda ascovirus 1a |
|
|
| Spodoptera frugiperda ascovirus 1a | [AM398843=NC_008361] | (SfAV-1a) |
| Spodoptera frugiperda ascovirus 1b |
| (SfAV-1b) |
| Spodoptera frugiperda ascovirus 1c | [AJ279824*] | (SfAV-1c) |
| Trichoplusia ni ascovirus 2a |
|
|
| Trichoplusia ni ascovirus 2a | [AJ312707*, AJ279826*] | (TnAV-2a) |
| Trichoplusia ni ascovirus 2b | [AJ279827*] |
|
| Diadromus pulcehllus ascovirus 4a** |
|
|
| Diadromus pulchellus ascovirus 4a | [CU469068=NC_011335] | (DpAV-4a) |
Species names are in italic script; names of strains are in roman script; synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
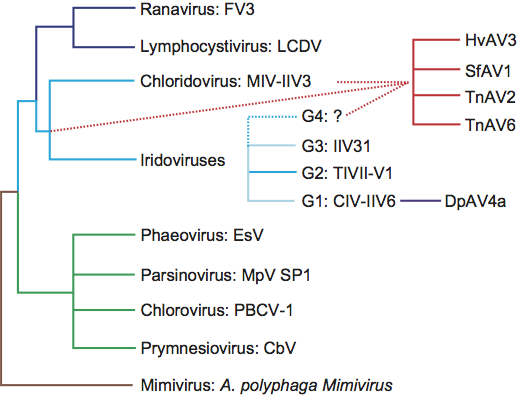
** Evidence obtained by Y. Bigot et al. (2009) shows that the species Diadromus pulcehllus ascovirus 4a does not belong to the same lineage as other ascoviruses and should be moved into either the, related, Iridoviridae family (see Figure 3) or an entirely new family.
List of other related viruses which may be members of the genus Ascovirus but have not been approved as species
| Helicoverpa armigera ascovirus 7a |
| (HaAV-7a) |
| Helicoverpa punctigera ascovirus 8a |
| (HpAV-8a) |
| Spodoptera exigua ascovirus 9a |
| (SeAV-9a) |
| Trichoplusia ni ascovirus 6a | [DQ517337=NC_008518] | (TnAV-6a) |
| (Trichoplusia ni ascovirus 2c) |
|
|
Phylogenetic relationships within the family
Phylogenetic relationships within the family have been inferred from the full length or partial peptide or DNA sequences of the DNA polymerase and major capsid protein. To date, the family appears to have evolved as a homogeneous lineage. Nevertheless, it must be noted that the current phylogenetic scheme is based on ascoviruses isolated from noctuid hosts. Whether ascoviruses occur in other lepidopteran or insect families in nature is unknown, but it is possible that such isolates could have dramatically different phylogenetic properties.
Phylogenetic relationships within the family Ascoviridae. Consensus neighbor-joining trees (PHYLIP) were constructed using alignments of ascovirus DNA polymerase and major capsid protein sequences. Trees were rooted using corresponding sequences from the Chilo iridescent virus type 6 (CIV) and DpAV4a. Bootstrap values are shown at branch nodes in the trees and branch lengths are proportional to genetic distances. In shaded boxes are located a lineage of ascovirus that were previously considered as different species but which, in the light of the most recent data, belong to a master species in which some features, such as the presence of occlusion bodies, can vary significantly.
Similarity with other taxa
Comparison of the gene content of three ascoviruses (HvAV-3e, SfAV-1a, TnAV-6a), DpAV-4a, two invertebrate iridoviruses (Chilo iridescent virus type [CIV] and mosquito iridovirus [MIV]) and eight vertebrate iridoviruses have demonstrated that these viruses share 28 core gene homologs and orthologs. The data also revealed that 67 core gene homologs are shared within the genus Ascovirus, with 29 and 42 of these being core genes in, respectively, invertebrate iridoviruses and DpAV4. Based on phylogenetic analyses of the 28 core genes, the following alternative theories for the origin of ascoviruses have been proposed:
- Ascoviruses and DpAV4 have different origins and their common molecular and biological features are due to the proximity of their ancestor and to convergences probably resulting from evolutionary processes occurring in similar environments.
- Ascoviridae and DpAV4 are two virus families that originated from invertebrate iridoviruses. Although the molecular clock varies significantly between core genes, Ascoviridae appear to have emerged early during the evolutionary radiation of invertebrate iridoviruses. Moreover, DpAV4 represents a more recent virus lineage that originated from a close iridovirus ancestor of Chilo iridescent virus (CIV, IIV6).
Further sequencing and phylogenetic analyses of invertebrate iridovirus genomes is required to definitively assess the origin of the Ascoviridae family among invertebrate iridoviruses.
Derivation of name
Asco: from the Greek for “sac”, referring to the virion-containing vesicles produced by cleavage of host cells, which are characteristic for all known viruses of this family.
Further reading
Asgari, S. (2006). Replication of Heliothis virescens ascovirus in insect cell lines. Arch. Virol., 61, 123-133.
Asgari, S., Davis, J., Wood, D., Wilson, P. and McGrath, A. (2007). Sequence and organization of the Heliothis virescens ascovirus genome. J. Gen. Virol., 88, 1120-1132.
Bideshi, D.K., Demattei, M.V., Rouleux-Bonnin, F., Stasiak, K., Tan, Y., Bigot, S., Bigot, Y. and Federici, B.A. (2006). Genomic sequence of Spodoptera frugiperda Ascovirus 1a, an enveloped, double-stranded DNA insect virus that manipulates apoptosis for viral reproduction. J. Virol., 80, 11791-11805.
Bideshi, D.K., Tan, Y., Bigot, Y. and Federici, B.A. (2005). A viral caspase contributes to modified apoptosis for virus transmission. Genes Dev., 19, 1416-1421.
Bigot, Y., Renault, S., Nicolas, J., Moundras, C., Demattei, M.V., Samain, S., Bideshi, D.K. and Federici, B.A. (2009). Symbiotic virus at the evolutionary intersection of three types of large DNA viruses; iridoviruses, ascoviruses, and ichnoviruses. PLoS One, 4, e6397.
Federici, B.A. (1983). Enveloped double-stranded DNA insect virus with novel structure and cytopathology. Proc. Natl Acad. Sci., USA, 80, 7664-7668.
Hussain, M. and Asgari, S. (2010). Functional analysis of a cellular microRNA in insect host–ascovirus interaction. J. Virol., 84, 612-620.
Tan, Y., Bideshi, D.K., Johnson, J.J., Bigot, Y. and Federici, B.A. (2009). Proteomic analysis of the Spodoptera frugiperda ascovirus 1a virion reveals 21 proteins. J. Gen. Virol., 90, 359-365.
Tan, Y., Spears, T., Bideshi, D.K., Johnson, J.J., Hice, R., Bigot, Y. and Federici, B.A. (2009). P64, a novel major virion DNA-binding protein potentially involved in condensing the Spodoptera frugiperda Ascovirus 1a genome. J. Virol., 83, 2708-2714.
Wang, L., Xue, J., Seaborn, C.P., Arif, B.M. and Cheng, X.W. (2006). Sequence and organization of the Trichoplusia ni ascovirus 2c (Ascoviridae) genome. Virology, 354, 167-177.
Contributed by
Bigot, Y., Asgari, S., Bideshi, D.K., Cheng X.W., Federici, B.A. and Renault, S.
Figures
Figures
Figure 1 Morphology of ascovirus virions. (A) Schematic illustration of the structure of a typical ascovirus virion. The virion consists of an inner particle and an outer envelope. The inner particle is complex and contains a DNA/protein core surrounded by an apparent unit membrane, the external surface of which bears a layer of distinctive protein subunits. (B,C) Respectively, ultrathin longitudinal- and cross-sections through typical ascovirus virions. The dense inner layer corresponds with the distinctive layer of subunits shown in top left. (D) Negatively stained preparations of virions from isolates of three different ascovirus species: top right, Spodoptera frugiperda ascovirus 1a (SfAV-1a); E, Trichoplusia ni ascovirus 2a (TnAV-2a), and (F), Heliothis virescens ascovirus 3a (HvAV-3a). The reticulate appearance of the virion is thought to be due to the superimposition of top and bottom layers of the inner particle and outer envelope. The bar represents 50 nm.
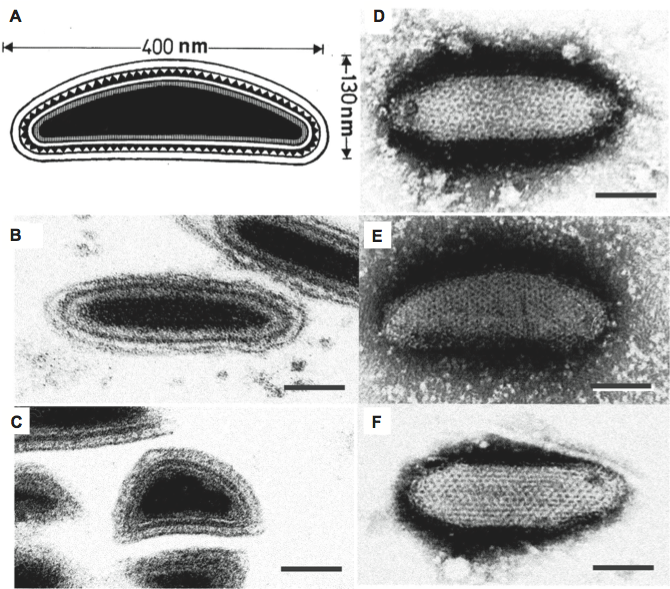
Figure 2 Phylogenetic relationships within the family Ascoviridae. Consensus neighbor-joining trees (PHYLIP) were constructed using alignments of ascovirus DNA polymerase and major capsid protein sequences. Trees were rooted using corresponding sequences from the Chilo iridescent virus type 6 (CIV) and DpAV4a. Bootstrap values are shown at branch nodes in the trees and branch lengths are proportional to genetic distances. In shaded boxes are located a lineage of ascovirus that were previously considered as different species but which, in the light of the most recent data, belong to a master species in which some features, such as the presence of occlusion bodies, can vary significantly.
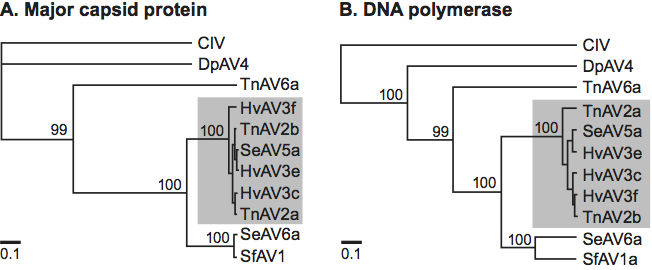
Figure 3 Synthesis of the evolutionary relationships among various genera of Mimiviridae (brown), Phycodnaviridae (green), Iridoviridae (invertebrate genera are in blue and vertebrate genera in dark blue), Ascoviridae (red) and DpAV4a (purple). The name of the principal virus representative of each genus is indicated. This synthetic tree was determined from results obtained with analyses of the 28 core genes. Plain lines represent verified relationships. Dotted lines indicate (putative) evolutionary pathways among virus families that require further support for confirmation. Phylogenetic analyses of major capsid protein (MCP) sequences revealed that at least three groups (G1, G2, G3) occurred within the iridovirus genus. A hypothetical fourth one, G4, was used to suggest the putative origin of the Ascoviridae.