Family: Adenoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Virions are non-enveloped, 70–90 nm in diameter. The icosahedral capsid consists of 240 non-vertex capsomers (hexons), 8–10 nm in diameter, and 12 vertex capsomers (penton bases), each with a fiber protruding from the virion surface giving the characteristic morphology (Figure 1). Penton base and fiber together make up the penton. The length of fibers examined so far ranges between 9 and 77.5 nm. Human adenoviruses 40 and 41 have fibers of two different lengths that occur alternately on the vertexes. Members of the genus Aviadenovirus have two fiber proteins per vertex. The 240 hexons are formed by the interaction of three identical polypeptides (designated II) and consist of two distinct parts: a triangular top with three “towers”, and a pseudohexagonal base with a central cavity. The hexon bases are tightly packed, forming a protein shell that protects the inner components. In members of the genus Mastadenovirus, 12 copies of polypeptide IX are found between nine hexons in the centre of each facet. Polypeptide IX is not present in the other four genera. Two monomers of IIIa are located underneath the vertex region. Multiple copies of protein VI form a ring underneath the peripentonal hexons. The 12 penton bases are each formed by the interaction of five polypeptides (III) and are tightly associated with one or two (only in aviadenoviruses) fibers, each consisting of three polypeptides (IV) that interact to form a shaft of characteristic length with a distal knob. The 12 pentons (III and IV) are less tightly associated with the neighbouring (peripentonal) hexons. Polypeptide VIII has been assigned to the inner surface of the hexon capsid. Other polypeptides (monomers of IIIa, trimers of IX and multimers of VI) are in contact with hexons, completing a continuous protein shell. Polypeptides VI and VIII appear to link the capsid to the virus core. The core consists of the DNA genome complexed with four polypeptides (V, VII, X, also known as mu or µ, and terminal protein). Protein V is found only in mastadenoviruses.
Physicochemical and physical properties
Virion Mr is 150–180×106; buoyant density in CsCl is 1.31–1.36 g cm−3. Viruses are stable on storage in the frozen state. They are stable to mild acid and insensitive to lipid solvents. Heat sensitivity varies in the different genera.
Nucleic acid
The genome is a single, linear molecule of dsDNA and contains an inverted terminal repetition (ITR). A virus-coded terminal protein (TP) is covalently linked to the 5′ end of each DNA strand. The size of genomes fully sequenced to date ranges between 26,163 and 48,395 bp, with ITRs of 36 to 371 bp. The G+C content of DNA varies between 33.6% and 66.9%. The central part of the genome is well conserved throughout the family, whereas the two ends show large variations in length and gene content (Figure 2).
Proteins
About 40 different polypeptides are produced, mostly via complex splicing mechanisms (Figure 2, Table 1). Almost a third compose the virion, including a virus-encoded cysteine protease (23 kDa), which is necessary for the processing of some precursor proteins (marked with p). With the exception of proteins V and IX, the other structural proteins are well conserved in every genus. Products of the four early regions (E1 to E4; E1 is often considered as two regions, E1A and E1B) facilitate extensive modulation of the host cell’s transcriptional machinery (E1 and E4), comprise the virus DNA replication complex (E2) and provide means for subverting host defense mechanisms (E3). E2 is well conserved throughout the family, while the length and gene content of E1, E3 and E4 show great variability even within genera. Intermediate (IX and IVa2) and late gene products (L1–L5) are concerned with virion assembly and maturation.
Table 1 Virus proteins as deduced from genome sequence of human adenovirus 2
|
kDa |
Transcription class |
Description |
Note |
|
13, 27, 32 |
E1A |
NS |
Only in mastadenoviruses |
|
16, 21 |
E1B |
NS |
Only in mastadenoviruses |
|
55 |
E1B |
NS |
Only in mastadenoviruses |
|
59 |
E2A |
NS; 72 kDa* DBP |
|
|
120 |
E2B |
NS; 140 kDa* DNA pol |
|
|
75 |
E2B |
S; Term, 87 kDa* pTP† |
|
|
4, 7, 8, 10, 12 |
E3 |
NS |
Only in mastadenoviruses |
|
13, 15, 15, 19 |
|
|
|
|
7, 13, 13, 14 |
E4 |
NS |
Only in mastadenoviruses |
|
15 |
E4 |
NS; 31 kDa* dUTPase |
Only in some mast- and aviadenoviruses |
|
17 |
E4 |
NS; 34 kDa* |
Only in mast- and atadenoviruses |
|
47 |
L1 |
NS; scaffolding 52/55 kDa* |
|
|
64 |
L1 |
S (pIIIa);† p-protein |
|
|
63 |
L2 |
S (III); penton base* |
|
|
22 |
L2 |
S (pVII);† major core |
|
|
42 |
L2 |
S (V); minor core |
Only in mastadenoviruses |
|
10 |
L2 |
S (pX);† X/µ |
|
|
27 |
L3 |
S (pVI)† |
|
|
109 |
L3 |
S (II); hexon |
|
|
23 |
L3 |
S; protease |
|
|
90 |
L4 |
NS; 100 kDa* |
|
|
25 |
L4 |
NS; 33 kDa* p-protein |
|
|
25 |
L4 |
S (pVIII)† |
|
|
62 |
L5 |
S (IV); fiber |
|
|
14 |
Intermediate |
S (IX) |
Only in mastadenoviruses |
|
51 |
Intermediate |
S (IVa2) |
|
Molecular masses are rounded to nearest 1000, and are presented as unmodified and uncleaved gene products. NS = non-structural; S = structural; p = precursor; p-protein = phosphoprotein; DBP = DNA-binding protein, DNA pol = DNA polymerase; TP = terminal protein; * = Mr values are significantly different from those obtained by SDS-PAGE; † = cleaved by viral protease.
Lipids
None reported.
Carbohydrates
Fiber proteins and some of the nonstructural proteins are glycosylated.
Genome organization and replication
Virus entry occurs by attachment via the fiber knob to different receptors on the surface of susceptible cells, and subsequent internalization via interaction between the penton base and cellular αv integrins. Protein VI mediates the release of virions from the endosomes, allowing dynein-mediated transport on microtubules to nuclear pores. After uncoating, the virus core is delivered to the nucleus, which is the site of virus RNA transcription, DNA replication and assembly. Virus infection mediates the early shut-off of host DNA synthesis and, later, synthesis of host mRNA and protein are also shut off. Transcription by host RNA polymerase II involves both DNA strands of the virus genome, and initiates (in human adenovirus 2, HAdV-2) from five early (E1A, E1B, E2, E3 and E4), two intermediate (IX and IVa2), the major late (L) and the U exon protein (UXP) late promoter in the pattern shown in Figure 3. All primary transcripts are capped and polyadenylated. There are complex splicing patterns to produce families of mRNAs. In primate adenoviruses, there are one or two virus-associated (VA) RNA genes, which are transcribed by cellular RNA polymerase III. These encode RNA products that facilitate translation of late mRNAs and blocking of the cellular interferon response. Similar VA RNA genes have not been identified in other adenoviruses. In some fowl adenoviruses, the existence of one VA RNA gene at a different genome position has been described, but these VA RNAs are not homologous to mastadenovirus VA RNAs.
Antigenic properties
Adenovirus serotypes are differentiated on the basis of neutralization assays. A serotype is defined as one that either exhibits no cross-reaction with others, or shows a homologous:heterologous titer ratio greater than 16 (in both directions). For homologous:heterologous titer ratios of 8 or 16, a serotype assignment is made if either the viral hemagglutinins are unrelated (as shown by lack of cross-reaction in hemagglutination-inhibition tests), or if substantial biophysical, biochemical or phylogenetic differences exist. Antigens at the surface of the virion are mainly type-specific. Hexons are involved in neutralization, and fibers in neutralization and hemagglutination-inhibition. Soluble antigens associated with virus infections include surplus capsid proteins that have not been assembled. As defined using monoclonal antibodies, hexons and other soluble antigens carry numerous epitopes that can be genus-, species- or type-specific. Free hexon protein reacts mainly as a genus-specific antigen. The genus-specific antigen is located on the basal surface of the hexon, whereas serotype-specific antigens are located mainly on the tower region.
Biological properties
The natural host range of adenovirus types is usually confined to a single species, or to closely related species. This also applies for cell cultures. Some human adenoviruses (HAdV) (mainly from members of the species Human adenovirus C) can cause productive infection in different animal cells (e.g. rodent or ruminant). Several human adenoviruses cause tumours in newborn hamsters. The majority of adenovirus infections in humans are subclinical. Direct or indirect transmission occurs from throat, faeces, eye or urine, depending on the virus type. Certain HAdV types (below in parentheses) are predominantly associated with specific pathology, such as adenoidal–pharyngeal conjunctivitis (3, 4, 7, 14), acute respiratory outbreaks (4, 7, 14, 21), epidemic keratoconjunctivitis (8, 19, 37, 53, 54) or venereal disease (37). HAdV-40 and HAdV-41 can be isolated in high yield from faeces of young children with acute gastroenteritis and are second only to rotaviruses as a major cause of infantile viral diarrhoea. HAdV-11, HAdV-34, and HAdV-35 cause persistent interstitial infection in the kidney and hemorrhagic cystitis, occurring most frequently in immuno-suppressed patients after organ transplantation. HAdV-42 to HAdV-51 were all isolated from AIDS patients. In other mammals, mastadenovirus infections are common, but manifest disease usually appears only if predisposing factors (e.g. management problems, crowding, shipping or concurrent bacterial infections) are present. Canine adenovirus (CAdV) seems to be an exception. CAdV-1 is the causative agent of infectious canine hepatitis (Rubarth disease), a life-threatening disease of puppies, and of encephalitis in numerous other carnivore species such as foxes, raccoons, bears and skunks. CAdV-2 causes infectious laryngotracheitis (kennel cough) in dogs, and this is common among breeder stocks. Adenoviruses infecting susceptible cells cause similar gross pathology, i.e. early rounding of cells and aggregation or lysis of chromatin, followed by the later appearance of characteristic basophilic or eosinophilic nuclear inclusions. HAdV-5 has been engineered and is used extensively as a gene vector. Other (including non-human) serotypes are being developed to overcome the problem posed by pre-existing neutralizing antibodies in the population, and also to achieve better targeting of specific organs and tissues.
Genus Mastadenovirus
Type species Human adenovirus C
Distinguishing features
Mastadenoviruses infect mammals only, and can be distinguished from members of other adenovirus genera traditionally by serology (genus members share complement-fixing antigen) and more recently (and preferably) by genome organization characteristics and phylogenetic distances. Virus infectivity is inactivated after heating at 56 °C for more than 10 min. Mastadenovirus genomes fully sequenced to date range between 30,536 bp (CAdV-1) and 37,860 bp (simian adenovirus 31.2; SAdV-31.2). The G+C content of the DNA varies between 43.6% (bovine adenovirus 2; BAdV-2) and 63.9% (porcine adenovirus 3; PAdV-3). The ITRs of mastadenoviruses are in general longer (93–371 bp) and more complex (containing a variety of cellular factor binding sites) than in members of the other genera. HAdV-2 comprises 35,937 bp and its ITR is 103 bp long.
Unique proteins of mastadenoviruses are proteins V and IX, and most of those coded by the E1A, E1B, E3 and E4 regions. As well as cementing the hexons on the outer surface of the capsid, protein IX also acts as a transcriptional activator and takes part in nuclear re-organization. Protein V is a core protein that, in association with cellular protein p32, seems to be involved in transport of viral DNA into the nucleus of the infected cell. The E3 and E4 proteins also often differ substantially between different mastadenovirus species.
Genome organization and replication have been most extensively studied for isolates of the species Human adenovirus C (Figure 3), and the findings seem to be generally applicable to all mastadenoviruses, except in the E3 and E4 regions. These early regions are also different in the non-primate mastadenoviruses. In the E4 region, a single homolog of the HAdV-2 34K protein exists in all mastadenoviruses and is duplicated in bovine adenovirus 3 and porcine adenovirus 5. The E3 region is also considerably shorter and less complex in the non-primate mastadenoviruses. The simplest E3 region, comprising a single gene, occurs in murine adenovirus 1 (MAdV-1) and MAdV-3.
Species demarcation criteria in the genus
Species demarcation is based on evolutionary distance as reflected by phylogenetic distances and genome organizational differences. The species contain similar types (designated by Arabic numbers) that were traditionally distinguished serologically (by virus neutralization). The serological type demarcation criterion is currently being replaced by criteria similar to those used for species demarcation. Species designation depends on several of the following characteristics:
- Phylogenetic distance (>5–15%, based primarily on distance matrix analysis of the DNA polymerase amino acid sequence)
- Genome organization (characteristically in the E3 region)
- Nucleotide composition (G+C%)
- Oncogenicity in rodents
- Host range
- Cross-neutralization
- Ability to recombine
- Number of VA RNA genes
- Hemagglutination
For example, if virus neutralization data are available, lack of cross-neutralization combined with a phylogenetic distance of more than 15% separates two types into different species. If the phylogenetic distance is less than 5%, any additional common grouping criteria from the list above may classify separate types into the same species even if they were isolated from different hosts. As an example, the most numerous types from the same host, the human adenoviruses, can be clearly separated into seven species supported by phylogenetic analysis, ability to recombine (e.g. between HAdV-1, 2, 5 and 6), growth characteristics (HAdV-40 and 41 show similar restricted capacity), oncogenicity and nucleotide composition (HAdV-12, 18 and 31, which are members of the species Human adenovirus A, share high oncogenicity in rodents and low G+C percentage in their genome). Adenoviruses isolated from chimpanzees resemble certain HAdVs to such an extent that they are classified into “human” adenovirus species. For example, simian adenoviruses (SAdVs) 22 to 25 belong to the species Human adenovirus E, and SAdV-21 belongs to the species Human adenovirus B.
List of species in the genus Mastadenovirus
|
Bovine adenovirus A |
|
|
|
Bovine adenovirus 1 |
[BD269513=NC_006324] |
(BAdV-1) |
|
Bovine adenovirus B |
|
|
|
Bovine adenovirus 3 |
[AF030154=AC_000002] |
(BAdV-3) |
|
Bovine adenovirus C |
|
|
|
Bovine adenovirus 10 |
[AF027599] |
(BAdV-10) |
|
Canine adenovirus |
|
|
|
Canine adenovirus 1 |
[Y07760=AC_000003] |
(CAdV-1) |
|
Canine adenovirus 2 |
[U77082=AC_000020] |
(CAdV-2) |
|
Equine adenovirus A |
|
|
|
Equine adenovirus 1 |
[L79955] |
(EAdV-1) |
|
Equine adenovirus B |
|
|
|
Equine adenovirus 2 |
[L80007] |
(EAdV-2) |
|
Human adenovirus A |
|
|
|
Human adenovirus 12 |
[X73487=NC_001460] |
(HAdV-12) |
|
Human adenovirus 18 |
[GU191019] |
(HAdV-18) |
|
Human adenovirus 31 |
[AM749299] |
(HAdV-31) |
|
Human adenovirus B |
|
|
|
Human adenovirus 3 |
[DQ086466=NC_011203] |
(HAdV-3) |
|
Human adenovirus 7 |
[AY495969=AC_000018] |
(HAdV-7) |
|
Human adenovirus 11 |
[AY163756=NC_011202] |
(HAdV-11) |
|
Human adenovirus 14 |
[AY803294] |
(HAdV-14) |
|
Human adenovirus 16 |
[AY601636] |
(HAdV-16) |
|
Human adenovirus 21 |
[AY601633] |
(HAdV-21) |
|
Human adenovirus 34 |
[AY737797] |
(HAdV-34) |
|
Human adenovirus 35 |
[AY271307] |
(HAdV-35) |
|
Human adenovirus 50 |
[AJ272612] |
(HAdV-50) |
|
Simian adenovirus 21 |
[AR101858=AC_000010] |
(SAdV-21) |
|
Human adenovirus C |
|
|
|
Bovine adenovirus 9 |
|
(BAdV-9) |
|
Human adenovirus 1 |
[AF534906] |
(HAdV-1) |
|
Human adenovirus 2 |
[J01917=NC_001405] |
(HAdV-2) |
|
Human adenovirus 5 |
[M73260=AC_000008] |
(HAdV-5) |
|
Human adenovirus 6 |
[HC492785] |
(HAdV-6) |
|
Simian adenovirus 31 |
[FJ025904] |
(SAdV-31) |
|
Human adenovirus D |
|
|
|
Human adenovirus 8 |
[AB448767] |
(HAdV-8) |
|
Human adenovirus 9 |
[AJ854486=NC_010956] |
(HAdV-9) |
|
Human adenovirus 10 |
[DQ149615] |
(HAdV-10) |
|
Human adenovirus 13 |
[DQ149616] |
(HAdV-13) |
|
Human adenovirus 15 |
[AB562586] |
(HAdV-15) |
|
Human adenovirus 17 |
[AF108105=AC_000006] |
(HAdV-17) |
|
Human adenovirus 19 |
[EF121005] |
(HAdV-19) |
|
Human adenovirus 20 |
[DQ149619] |
(HAdV-20) |
|
Human adenovirus 22 |
[FJ404771] |
(HAdV-22) |
|
Human adenovirus 23 |
[DQ149621] |
(HAdV-23) |
|
Human adenovirus 24 |
[DQ149622] |
(HAdV-24) |
|
Human adenovirus 25 |
[DQ149623] |
(HAdV-25) |
|
Human adenovirus 26 |
[EF153474] |
(HAdV-26) |
|
Human adenovirus 27 |
[DQ149625] |
(HAdV-27) |
|
Human adenovirus 28 |
[FJ824826] |
(HAdV-28) |
|
Human adenovirus 29 |
[AB562587] |
(HAdV-29) |
|
Human adenovirus 30 |
[DQ149628] |
(HAdV-30) |
|
Human adenovirus 32 |
[DQ149629] |
(HAdV-32) |
|
Human adenovirus 33 |
[DQ149630] |
(HAdV-33) |
|
Human adenovirus 36 |
[GQ384080] |
(HAdV-36) |
|
Human adenovirus 37 |
[DQ900900] |
(HAdV-37) |
|
Human adenovirus 38 |
[DQ149633] |
(HAdV-38) |
|
Human adenovirus 39 |
[DQ149634] |
(HAdV-39) |
|
Human adenovirus 42 |
[DQ149635] |
(HAdV-42) |
|
Human adenovirus 43 |
[DQ149636] |
(HAdV-43) |
|
Human adenovirus 44 |
[DQ149637] |
(HAdV-44) |
|
Human adenovirus 45 |
[DQ149638] |
(HAdV-45) |
|
Human adenovirus 46 |
[AY875648] |
(HAdV-46) |
|
Human adenovirus 47 |
[DQ149640] |
(HAdV-47) |
|
Human adenovirus 48 |
[EF153473] |
(HAdV-48) |
|
Human adenovirus 49 |
[DQ393829] |
(HAdV-49) |
|
Human adenovirus 51 |
[DQ149642] |
(HAdV-51) |
|
Human adenovirus 53 |
[FJ169625] |
(HAdV-53) |
|
Human adenovirus 54 |
[AB333801=NC_012959] |
(HAdV-54) |
|
Human adenovirus E |
|
|
|
Human adenovirus 4 |
[AY487947=NC_003266] |
(HAdV-4) |
|
Simian adenovirus 22 |
[AY530876] |
(SAdV-22) |
|
Simian adenovirus 23 |
[AY530877] |
(SAdV-23) |
|
Simian adenovirus 24 |
[AY530878] |
(SAdV-24) |
|
Simian adenovirus 25 |
[AR101859=AC_000011] |
(SAdV-25) |
|
Human adenovirus F |
|
|
|
Human adenovirus 40 |
[L19443=NC_001454] |
(HAdV-40) |
|
Human adenovirus 41 |
[DQ315364] |
(HAdV-41) |
|
Human adenovirus G |
|
|
|
Human adenovirus 52 |
[DQ923122] |
(HAdV-52) |
|
Simian adenovirus 1 |
[AY771780=NC_006879] |
(SAdV-1) |
|
Simian adenovirus 2 |
[SAU03008] |
(SAdV-2) |
|
Simian adenovirus 7 |
[DQ792570] |
(SAdV-7) |
|
Simian adenovirus 11 |
[SAU03014] |
(SAdV-11) |
|
Simian adenovirus 12 |
|
(SAdV-12) |
|
Simian adenovirus 15 |
[SAU03016] |
(SAdV-15) |
|
Murine adenovirus A |
|
|
|
Murine adenovirus 1 |
[NC_000942] |
(MAdV-1) |
|
Murine adenovirus C |
|
|
|
Murine adenovirus 3 |
[EU835513=NC_012584] |
(MAdV-3) |
|
Ovine adenovirus A |
|
|
|
Bovine adenovirus 2 |
[AF252854=AC_000001] |
(BAdV-2) |
|
Ovine adenovirus 2 |
[DQ630755] |
(OAdV-2) |
|
Ovine adenovirus 3 |
[DQ630756] |
(OAdV-3) |
|
Ovine adenovirus 4 |
[DQ630757] |
(OAdV-4) |
|
Ovine adenovirus 5 |
[DQ630758] |
(OAdV-5) |
|
Ovine adenovirus B |
|
|
|
Goat adenovirus 2 |
[DQ630760] |
(GAdV-2) |
|
Ovine adenovirus 1 |
[DQ630754] |
(OAdV-1) |
|
Porcine adenovirus A |
|
|
|
Porcine adenovirus 1 |
[L43364] |
(PAdV-1) |
|
Porcine adenovirus 2 |
[L43365] |
(PAdV-2) |
|
Porcine adenovirus 3 |
[AF083132=NC_005869] |
(PAdV-3) |
|
Porcine adenovirus B |
|
|
|
Porcine adenovirus 4 |
[U13893] |
(PAdV-4) |
|
Porcine adenovirus C |
|
|
|
Porcine adenovirus 5 |
[AF289262=NC_002702] |
(PAdV-5) |
|
Simian adenovirus A |
|
|
|
Simian adenovirus 3 |
[AY598782=NC_006144] |
(SAdV-3) |
|
Simian adenovirus 4 |
[SAU03010] |
(SAdV-4) |
|
Simian adenovirus 6 |
[CQ982401] |
(SAdV-6) |
|
Simian adenovirus 9 |
[SAU03012] |
(SAdV-9) |
|
Simian adenovirus 10 |
[SAU03013] |
(SAdV-10) |
|
Simian adenovirus 14 |
[SAU03016] |
(SAdV-14) |
|
Simian adenovirus 48 |
[HQ241818] |
(SAdV-48) |
|
Tree shrew adenovirus |
|
|
|
Tree shrew adenovirus 1 |
[AF258784=NC_004453] |
(TSAdV-1) |
Species names are in italic script; names of types are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed. Full genome sequences are available from 19 further novel chimpanzee, eight gorilla and six bonobo adenoviruses. These belong to the species Human adenovirus B, C and E, with representatives in each. Due to lack of space and confirmation of their type demarcation, they are not listed.
List of other related viruses which may be members of the genus Mastadenovirus but have not been approved as species
|
Alpaca adenovirus 1 |
[GQ499375] |
(AlAdV-1) |
|
Bat adenovirus 1 (FBV1) |
[AB303301] |
(BtAdV-1) |
|
Bat adenovirus 2 (PPV1) |
[FJ983127] |
(BtAdV-2) |
|
Bat adenovirus 3 (TJM) |
[GU226970] |
(BtAdV-3) |
|
Guinea pig adenovirus 1 |
[X95630] |
(GPAdV-1) |
|
Murine adenovirus 2 |
[HM049560=NC_014899] |
(MAdV-2) |
|
Ovine adenovirus 6 |
[DQ630759] |
(OAdV-6) |
|
Simian adenovirus 5, 8, 13, 16–20 |
[18: FJ025931] |
(SAdV-5, 8, 13, 16–20) |
|
Squirrel adenovirus 1 |
[GU735084] |
(SqAdV-1) |
Genus Aviadenovirus
Type species Fowl adenovirus A
Distinguishing features
Aviadenoviruses are serologically distinct from members of the other adenovirus genera and they only infect birds. The virions contain two fibers per vertex. Fowl adenovirus 1 (FAdV-1), FAdV-4 and turkey adcnovirus 1 (TAdV-1) have two fiber genes, and two projections (in case of FAdV-1, of considerably different lengths) on each penton base. Other FAdVs also have two fibers per vertex, but apparently only one fiber gene, and the fiber shafts are of similar lengths. The long fiber of FAdV-1 uses the coxsackievirus and adenovirus receptor (CAR) for attachment to the cell.
Aviadenovirus genomes are considerably larger (20–45%) than those of mastadenoviruses. Five aviadenovirus [FAdV-1, FAdV-4, “FAdV-8”, FAdV-9 and TAdV-1] genomes have been fully sequenced, and range between 43,804 bp (FAdV-1) and 45,667 (FAdV-4). These are thought to represent the longest adenovirus DNA molecules after that of white sturgeon adenovirus. The G+C content of partial or complete sequences of aviadenovirus genomes varies between 53.8 and 66.9%. The size of sequenced aviadenovirus ITRs are between 54 and 95 bp long. The genomic organization of aviadenoviruses is also different from that of adenoviruses in other genera (Figure 2). The genes of proteins V and IX, as well as genes in mastadenovirus early regions E1 and E3, are missing. The E4 region may be translocated from its position in mastadenovirus genomes, resulting in the gene encoding dUTP pyrophosphatase (dUTPase, not present in every mastadenovirus) being on the left end of the genome, rather than the right end. (Alternatively, this gene may have been captured independently by ancestors of aviadenoviruses and mastadenoviruses.) The organization of the central part of the genome containing the late genes and the E2 region is similar to that of mastadenoviruses. The right end of the genome contains several transcription units, which are unique to aviadenoviruses. The majority of genes and proteins from this region have not yet been characterized in detail. A novel protein GAM-1 of FAdV-1 has been demonstrated to have an anti-apoptotic effect, and to activate the heat-shock response in the infected cell. GAM-1, in synergy with another novel protein encoded by ORF22, binds the retinoblastoma protein and can activate the E2F pathway. Additional, and as yet uncharacterized predicted gene products, exhibit sequence homology to proteins of other viruses, such as the non-structural protein NS1 (also known as Rep) of parvoviruses, and a triacylglycerol lipase, a homolog of which also occurs in an avian herpesvirus (Marek’s disease virus). Aviadenoviruses possess no complement-fixing antigen in common with the members of the other genera. There are isolates where serum neutralization cannot differentiate clearly between the serotypes. The introduction of FAdV-8a and 8b was deemed necessary because of the inconsistency in the type-numbering scheme used in different countries and continents over the years. Avian adenoviruses have been associated with diverse disease patterns, including inclusion body hepatitis, bronchitis, pulmonary congestion and oedema in different bird species. Hydropericardium syndrome is caused by FAdV-4 in chickens, mainly in Asia. Falcon adenovirus 1 has caused fatalities in different falcon species. FAdV-1 (CELO virus), 9 and 10 are being studied for their feasibility as gene delivery vectors.
Species demarcation criteria in the genus
Species designation depends on several of the following characteristics:
- Phylogenetic distance (>5–15%, based primarily on distance matrix analysis of the DNA polymerase amino acid sequence)
- Genome organization (characteristically in the region at the right end of the genome)
- RFLP analysis
- Host range
- Pathogenicity
- Cross-neutralization
- Ability to recombine
For example, the fowl adenovirus serotypes can be grouped into five species on the basis of phylogeny, genome organization, RFLP profiles and the lack of significant cross-neutralization.
List of species in the genus Aviadenovirus
Note: A specific problem that has been addressed but not resolved is the lack of consensus in the numbering of the individual fowl adenovirus serotypes. Strains deposited in the American Type Culture Collection are numbered inconsistently in relation to the majority of newer publications. For this reason, one representative strain of each serotype is also listed (in parentheses).
|
Falcon adenovirus A |
|
|
|
Falcon adenovirus 1 |
[AY683541] |
(FaAdV-1) |
|
Fowl adenovirus A |
|
|
|
Fowl adenovirus 1 (CELO) |
[U46933=AC_000014] |
(FAdV-1) |
|
Fowl adenovirus B |
|
|
|
Fowl adenovirus 5 (340) |
[AF508952] |
(FAdV-5) |
|
Fowl adenovirus C |
|
|
|
Fowl adenovirus 4 (ON1) |
[GU188428=NC_015323] |
(FAdV-4) |
|
Fowl adenovirus 10 (CFA20) |
[AF160185] |
(FAdV-10) |
|
Fowl adenovirus D |
|
|
|
Fowl adenovirus 2 (P7-A) |
[AF339915] |
(FAdV-2) |
|
Fowl adenovirus 3 (75) |
[AF508949] |
(FAdV-3) |
|
Fowl adenovirus 9 (A2-A) |
[AF083975=AC_000013] |
(FAdV-9) |
|
Fowl adenovirus 11 (380) |
[AF339925] |
(FAdV-11) |
|
Fowl adenovirus E |
|
|
|
Fowl adenovirus 6 (CR119) |
[AF508954] |
(FAdV-6) |
|
Fowl adenovirus 7 (YR36) |
[AF508955] |
(FAdV-7) |
|
Fowl adenovirus 8a (CFA40) |
[AF155911] |
(FAdV-8a) |
|
Fowl adenovirus 8b (764) |
[AF508958] |
(FAdV-8b) |
|
Goose adenovirus |
|
|
|
Goose adenovirus 1 |
|
(GoAdV-1) |
Species names are in italic script; names of types and isolates ( ) are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Aviadenovirus but have not been approved as species
|
Duck adenovirus 2 |
|
(DAdV-2) |
|
Meyer’s parrot adenovirus 1 |
[AY644731] |
|
|
Pigeon adenovirus 1 |
[FN824512] |
(PiAdV-1) |
|
Psittacine adenovirus 1 |
[EF442329] |
(PsAdV-1) |
|
Turkey adenovirus 1 |
[GU936707= NC_014564] |
(TAdV-1) |
|
Turkey adenovirus 2 |
[GU936708] |
(TAdV-2) |
Genus Atadenovirus
Type species Ovine adenovirus D
Distinguishing features
Atadenoviruses are serologically distinct from viruses in the other adenovirus genera, and their genomic organization and capsid protein complement also differ. Atadenoviruses have been detected in a broad range of hosts, including scaled reptiles (order Squamata) and species ranging from birds to ruminants and a marsupial. Virions are relatively heat stable and retain substantial infectivity after treatment for 30 min at 56 °C, conditions that inactivate mastadenovirions. The genome size of sequenced isolates ranges from 29,576 (ovine adenovirus 7, OAdV-7) to 33,213 bp (duck adenovirus 1, DAdV-1) with ITRs of 46 (OAdV-7) to 118 bp (snake adenovirus 1, SnAdV-1). For ruminant, marsupial and avian atadenoviruses, the G+C content of the DNA is low and varies between 33.6 (OAdV-7) and 43.0% (DAdV-1). The corresponding high A+T content was deemed sufficiently characteristic to be used as the basis of the name of the genus, though atadenoviruses originating from scaled reptiles have a non-biased nucleotide composition (50.0% G+C in SnAdV-1). The proteins encoded by an atadenovirus genome are summarized in Table 2. The central part of the atadenovirus genome is similar to that of mastadenoviruses (except that there are no protein V and IX genes), whereas the extremities differ markedly. Atadenoviruses have several unique proteins, including some that may be distant homologs of proteins in other adenovirus genera. The left end of the genome carries a gene for p32K, a unique structural protein. At this end, gene LH1 is also unique to the genus but not present in all members. The right end of the genome contains genes that are related to each other, suggestive of gene duplication. There are two E4 34K gene homologs (E4.2 and E4.3), and one to five gene RH homologs. At this end, genes E4.1 and RH1–6 are also unique to the genus but are not present in all members. The proteins encoded by genes LH3 and E4.3 (and its paralog, E4.2) show limited similarity to mastadenovirus proteins E1B 55K and E4 34K, respectively. However, LH3 is a structural protein that forms prominent “knobs” on the virion surface that distinguish atadenoviruses structurally from other known AdVs (Figure 4). LH3 is located in the same relative position among the hexon subunits as protein IX (present only in mastadenoviruses) but sits on top of the “central” (so called H3) hexon trimers, appearing to hold the outer capsid together. Particles that lack the LH3 or p32K proteins are viable although less stable. (Mastadenoviruses that lack proteins V or IX are also viable.) No immunomodulatory genes such as those found in the mastadenovirus E3 region have yet been identified. DAdV-1 has a unique genome region at the far right end with seven uncharacterized ORFs that may be host-specific in function. ORF5 and ORF6 are related to each other. This unique region of DAdV-1 also contains a VA RNA gene that is supposedly homologous to that of FAdV-1.
Certain atadenoviruses can cause hemorrhagic epizooty in free-living ruminants. DAdV-1 is also associated with a specific disease of hens that is characterized globally by sharp decreases in egg production (egg drop syndrome). Due to the lack of pre-existing immunity in humans and its bio-safety profile, OAdV-7 has been developed as a gene delivery vector intended for human vaccine and gene therapy applications.
Table 2 Virus proteins as deduced from the genome sequence of ovine adenovirus 7
|
kDa |
Transcription class |
Description |
Note |
|
32 |
Timing not known |
S (p32K†) |
Unique to atadenoviruses |
|
13 |
LH1 |
NS |
ORF not in DAdV-1 |
|
14.7 |
LH2 |
NS |
|
|
42.8 |
LH3 |
S |
Distant homolog of mastadenovirus |
|
|
Timing not known |
|
E1B 55K |
|
43 |
E2 |
NS; DBP |
|
|
123 |
E2 |
NS; DNA pol |
|
|
67.1 |
E2 |
S; Term, pTP† |
|
|
12.9, 20.9, 19.8, 19.8 |
RH1, RH2, RH4, RH6, early |
NS |
F-box proteins unique for atadenovirusesHomology to each other |
|
22.6 |
RH5, early |
NS |
ORF not in DAdV-1 |
|
17.1 |
E4.1, early |
NS |
ORF not in DAdV-1 |
|
25.6, 30.8 |
E4.2, E4.3, early |
NS |
Distant homologs of mastadenovirus E4 34K |
|
38.2 |
Early and late |
NS; scaffolding |
|
|
|
|
52/55 kDa* |
|
|
58.4 |
Late |
S (pIIIa);† p-protein |
|
|
51 |
Late |
S (III); penton |
|
|
|
|
base* |
Lacks integrin binding motif |
|
12.9 |
Late |
S (pVII);† major core |
|
|
7.3 |
Late |
S (pX);† X/µ |
|
|
24.5 |
Late |
S (pVI)† |
|
|
102 |
Late |
S (II); hexon |
|
|
23 |
Late |
S; protease |
|
|
72 |
Late |
NS; 100 kDa* |
Hexon assembly protein |
|
15.7 |
Late |
NS; 33 kDa* |
p-protein not found in DAdV-1 |
|
24.7 |
Late |
S (pVIII)† |
|
|
58.2 |
Late |
S (IV); fiber |
Cell attachment protein |
|
37.5 |
Timing not known |
S (IVa2) |
|
Molecular masses are presented as unmodified and uncleaved gene products. S = structural; NS = non-structural; LH = left-hand end [genes]; RH = right-hand end [genes]; p = precursor; p-protein = phosphoprotein; DBP = DNA-binding protein; DNA pol = DNA polymerase; Term = terminal protein; * = Mr values are significantly different from those obtained by SDS-PAGE; † = cleaved by viral protease. All NS proteins are hypothetical until characterized.
Species demarcation criteria in the genus
Species designation depends on several of the following characteristics:
- Phylogenetic distance (>5–15%, based primarily on distance matrix analysis of the DNA polymerase amino acid sequence)
- Host range
- DNA hybridization
- Nucleotide composition (G+C%)
- Cross-neutralization
- Gene organization of the right end of the genome
List of species in the genus Atadenovirus
|
Bovine adenovirus D |
|
|
|
Bovine adenovirus 4 |
[AF036092=NC_002685] |
(BAdV-4) |
|
Bovine adenovirus 5 |
[AF207658] |
(BAdV-5) |
|
Bovine adenovirus 8 |
[AF238233] |
(BAdV-8) |
|
Bovine adenovirus strain Rus |
[AF238880] |
(BAdV-Rus) |
|
Duck adenovirus A |
|
|
|
Duck adenovirus 1 |
[Y09598=AC_000004] |
(DAdV-1) |
|
Ovine adenovirus D |
|
|
|
Goat adenovirus 1 |
[AF207660] |
(GAdV-1) |
|
Ovine adenovirus 7 |
[U40839=NC_004037] |
(OAdV-7) |
|
Possum adenovirus |
|
|
|
Possum adenovirus 1 |
[AF338822] |
(PoAdV-1) |
|
Snake adenovirus A |
|
|
|
Snake adenovirus 1 |
[DQ106414=NC_009989] |
(SnAdV-1) |
Species names are in italic script; names of types and isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Atadenovirus but have not been approved as species
|
Asp viper adenovirus |
[EU914209] |
|
|
Bearded dragon adenovirus 1 (agamid adenovirus 1) |
[AY576678] |
(BDAdV-1) |
|
Blue-tongued skink adenovirus |
[AY576682] |
|
|
Bovine adenovirus 6 |
[AF207659] |
(BAdV-6) |
|
Bovine adenovirus 7 |
[AF238232] |
(BAdV-7) |
|
Chameleon adenovirus 1 |
[AY576679] |
(ChAdV-1) |
|
Emerald monitor adenovirus |
[EU914208] |
|
|
Gecko adenovirus 1 |
[AY576677] |
(GeAdV-1) |
|
Gila adenovirus 1 |
[AY576680] |
|
|
Hagen’s pit viper adenovirus |
[EU851415] |
|
|
Mexican beaded lizard adenovirus 1 |
[EU914207] |
|
|
Odocoileus adenovirus 1 |
[AF198354] |
(OdAdV-1) |
|
Snake adenovirus 2 |
[FJ012163] |
(SnAdV-2) |
|
Snake adenovirus 3 |
[FJ012164] |
(SnAdV-3) |
|
Tokay gecko adenovirus |
[AY576681] |
|
Genus Siadenovirus
Type species Frog adenovirus
Distinguishing features
Siadenoviruses are serologically and phylogenetically distinct from members of the other adenovirus genera. This genus comprises only three accepted members, frog adenovirus 1 (FrAdV-1), turkey adenovirus 3 (TAdV-3) and raptor adenovirus 1 (RAdV-1). FrAdV-1 was isolated from an amphibian (frog) and TAdV-3 from birds (turkey, pheasant and chicken). RAdV-1 was detected in captive raptors by PCR. The RAdV-1 genome was the first adenovirus genome to be fully sequenced solely by PCR-based methods without virus isolation. The genomes of the three siadenovirus types represent the shortest adenovirus genomes known to date. Their lengths are between 26,163 and 26,283 bp, with G+C contents of 34.9 to 38.5%, and ITRs of 29–39 bp. The genomic organization of siadenoviruses is also characteristic and different from that of other genera. The genes of proteins V and IX, and homologs in mastadenovirus early regions E1, E3 and E4 are absent. Apart from the proteins conserved in all adenoviruses, there are only five ORFs potentially encoding novel proteins. At the left end of the genome, the first putative gene encodes a protein that is related to sialidases. Adjacent to it is a novel ORF predicted to code for a highly hydrophobic protein. The gene named “E3” solely because of its position between the pVIII and fiber genes is not homologous to any of the mastadenovirus E3 genes (or to any other known genes). The right end of the genome harbours ORF7 and ORF8. TAdV-3 has no common complement-fixing antigen with other adenoviruses isolated from birds and classified in the aviadenovirus or atadenovirus genera. FrAdV-1 is supposedly non-pathogenic. TAdV-3 is associated with specific diseases in different hosts (hemorrhagic enteritis in turkey, marble spleen disease in pheasants and splenomegaly in chickens).
Species demarcation criteria in the genus
Species designation depends on the following characteristics:
- Phylogenetic distance (>5–15%, based primarily on distance matrix analysis of the DNA polymerase amino acid sequence)
- Host range
List of species in the genus Siadenovirus
|
Frog adenovirus |
|
|
|
Frog adenovirus 1 |
[AF224336=NC_002501] |
(FrAdV-1) |
|
Raptor adenovirus A |
|
|
|
Raptor adenovirus 1 |
[EU715130=NC_015455] |
(RAdV-1) |
|
Turkey adenovirus A |
|
|
|
Turkey adenovirus 3 |
[AF074946=AC_000016] |
(TAdV-3) |
Species names are in italic script; names of types are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Siadenovirus but have not been approved as species
|
Budgerigar adenovirus 1 |
[AB485763] |
|
|
Great tit adenovirus 1 |
[FJ849795] |
(GTAdV-1) |
|
adenovirus 2 |
[EU056825] |
(PsAdV-2) |
|
Sulawesi tortoise adenovirus 1 |
[EU056826] |
(STAdV-1) |
Genus Ichtadenovirus
Type species Sturgeon adenovirus A
Distinguishing features
The single known member of this genus, white sturgeon adenovirus 1 (WSAdV-1), is the only confirmed fish adenovirus. While the host is very divergent from those of other AdVs, phylogenetic calculations and a unique genome organization further distinguish this virus from all other adenoviruses. The WSAdV-1 genome is 48,395 bp, and thus it is longer than the genome of any known adenovirus. The G+C content is 42.7%. The fiber gene was not found in its usual position towards the right end of the genome, but fiber gene homologs were discovered at the left end. The virus seems to be non-pathogenic for fish.
List of species in the genus Ichtadenovirus
|
Sturgeon adenovirus A |
|
|
|
White sturgeon adenovirus 1 |
[AJ495768] |
(WSAdV-1) |
Species names are in italic script; names of types are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Ichtadenovirus but have not been approved as species
None reported.
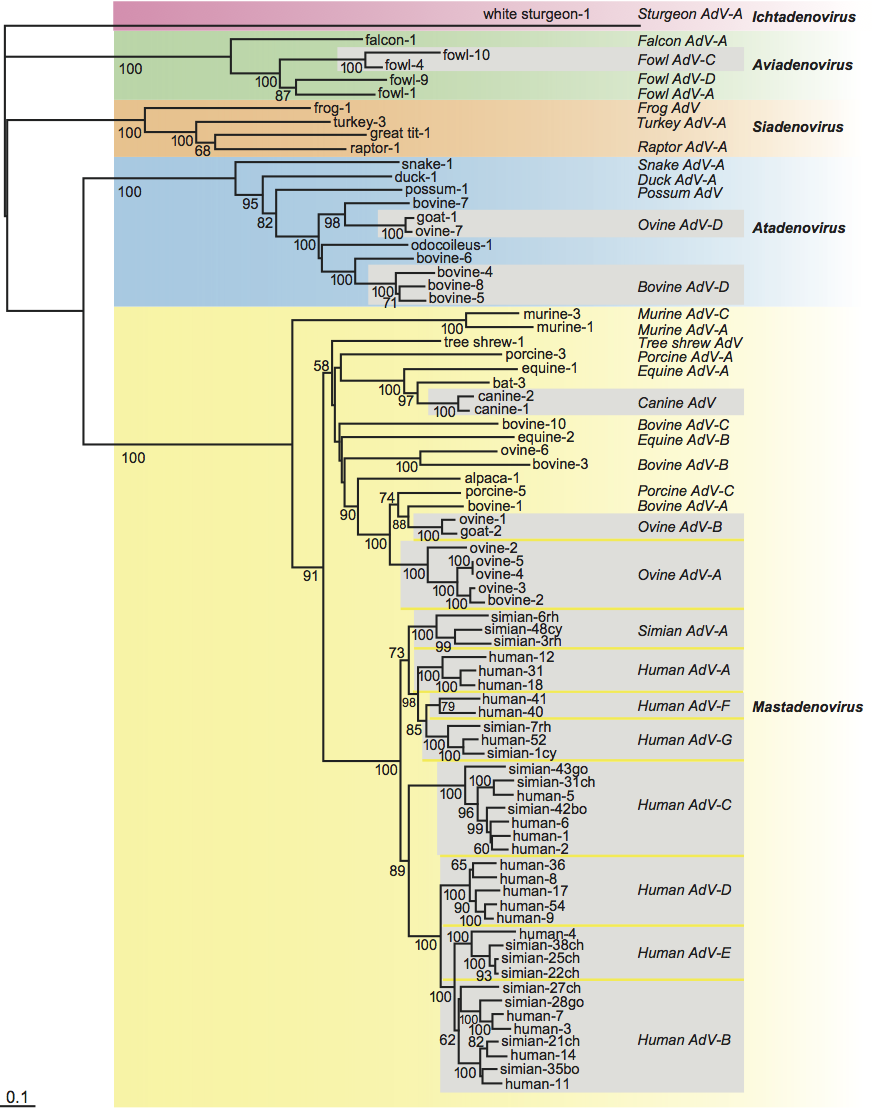
Phylogenetic relationships within the family
Consistent with specific characteristics of genome organization, phylogenetic calculations result in clear separation of five different clusters corresponding to the five genera (Mastadenovirus, Aviadenovirus, Atadenovirus, Siadenovirus and Ichtadenovirus; Figure 5). Within the genera, evolutionary distances among adenoviruses resemble those of their host. There are some exceptions, where very distantly related viruses infect the same host. Adenovirus types isolated from cattle, sheep or chicken appear on very distant branches, and even in separate clusters corresponding to different genera. Beside the co-evolution that has been hypothesized for animals and their adenoviruses, multiple host switches may also have occurred. For example, adenoviruses from scaled reptiles switched to ruminants and birds.
Similarity with other taxa
A dsDNA bacteriophage, PRD1 (in the family Tectiviridae), shares a similar virion architecture (an icosahedral capsid with fiber-like projections) with adenoviruses. The 15 kbp genome of PRD1 has ITRs and contains genes encoding terminal protein and DNA polymerase arranged in the same order as in adenoviruses. The terminal protein also acts as primer in PRD1 DNA replication. A study on the resolution of the main capsid proteins (P3 of PRD1 and hexon of HAdV-2) revealed a very similar arrangement and structure, and again suggested an evolutionary link between the two viruses. Fungi and plants also have a linear plasmid (killer plasmid in yeast) either in the cytoplasm or in the mitochondria that has adenovirus-like features (an ITR and a terminal protein gene adjacent to a DNA polymerase gene). Sulfolobus turreted icosahedral virus, which infects a crenarchaeal host (in domain Archaea), and also two archaeal proviruses (TKV4 and MVV), which are integrated into the 5′- and 3′-distal regions of tRNA genes of the euryarchaeal species Thermococcus kodakaraensis KOD1 and Methanococcus voltae A3, respectively, could also be placed into the PRD1-adenovirus lineage based on established or predicted three-dimensional structures of their major capsid protein and on sharing a characteristic ATPase.
Adenoviruses also show some similarity to other viruses. The fibers of many adenovirus types use the same cellular receptor (CAR) for attachment as coxsackie B viruses. Adenovirus fibers were reported to show structural similarity with reovirus attachment protein sigma 1, which binds the JAM (junction adhesion molecule) receptor. Adenoviruses may occur together with dependent parvoviruses, for which they may provide helper functions. Similarity was observed between certain proteins of the E3 region of human adenoviruses and the RL11 gene family of human cytomegalovirus (a herpesvirus). The primary structure of the p32K protein, which is characteristic of atadenoviruses, has similarity to bacterial small acid soluble proteins (SASPs) commonly found in various spore-forming bacteria.
Derivation of names
Adeno: from Greek aden, adenos, “gland”; in recognition of the fact that adenoviruses were first isolated from human adenoid tissue.
At: from adenine and thymine, in recognition that the genome of the first recognized members of the genus (from ruminant, avian and marsupial hosts) has a remarkably high A+content.
Avi: from Latin avis, “bird”.
Icht: from Greek ichthys, “fish”.
Mast: from Greek mastos, “breast”.
Si: from sialidase, in recognition that members of the genus have a putative sialidase homolog.
Further reading
Benk, M. (2008). Adenoviruses. Pathogenesis. In: B.W.J. Mahy and M.H.V. van Regenmortel (Eds.), Encyclopedia of Virology, 3rd edition Elsevier, Oxford, vol. 1, pp. 24-29.
Corredor, J.C., Garceac, A., Krell, P.J. and Nagy, É. (2008). Sequence comparison of the right end of fowl adenovirus genomes. Virus Genes, 36, 331-344.
Davison, A.J., Benk, M., Harrach, B. (2003). Genetic content and evolution of adenoviruses. J. Gen. Virol., 84, 2895-2908.
Farkas, S.L., Harrach, B. and Benk, M. (2008). Completion of the genome analysis of snake adenovirus type 1, a representative of the reptilian lineage within the novel genus Atadenovirus. Virus Res., 132, 132-139.
Harrach, B. (2008). Adenoviruses. General features. In: B.W.J. Mahy and M.H.V. van Regenmortel (Eds.), Encyclopedia of Virology, 3rd edn. Elsevier, Oxford, vol. 1, pp. 1-9.
Jones, M.S. 2nd, Harrach, B., Ganac, R.D., Gozum, M.M., Dela Cruz, W.P., Riedel, B., Pan, C., Delwart, E.L. and Schnurr, D.P. (2007). New adenovirus species found in a patient presenting with gastroenteritis. J. Virol., 81, 5978-5984.
Kovács, E.R., Jánoska, M., Dán, Á., Harrach, B. and Benk, M. (2010). Recognition and partial genome characterization by non-specific DNA amplification and PCR of a new siadenovirus species in a sample originating from Parus major, a great tit. J. Virol. Methods, 163, 262-268.
Pantelic, R.S., Lockett, L.J., Rothnagel, R., Hankamer, B. and Both, G.W. (2008). Cryoelectron microscopy map of Atadenovirus reveals cross-genus structural differences from human adenovirus. J. Virol., 82, 7346-7356.
Roy, S., Vandenberghe, L.H., Kryazhimskiy, S., Grant, R., Calcedo, R., Yuan, X., Keough, M., Sandhu, A., Wang, Q., Medina-Jaszek, C.A., Plotkin, J.B. and Wilson, J.M. (2009). Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog., 5, e1000503.
Wellehan, J.F.X., Johnson, A.J., Harrach, B., Benk, M., Pessier, A.P., Johnson, C.M., Garner, M.M., Childress, A. and Jacobson, E.R. (2004). Detection and analysis of six lizard adenoviruses by consensus primer PCR provides further evidence of a reptilian origin for the atadenoviruses. J. Virol., 78, 13366-13369.
Contributed by
Harrach, B., Benkö, M., Both, G.W., Brown, M., Davison, A.J., Echavarría, M., Hess, M., Jones M.S., Kajon, A., Lehmkuhl, H.D., Mautner, V., Mittal, S.K. and Wadell, G.
Figures
Figure 1 (Left) Cryo-EM reconstruction of a human adenovirus 2 particle (Stewart, P.L. et al. (1991). Cell, 67, 145154). (Centre) Stylized section of a mastadenovirus particle. For a description of the capsid proteins (II, III, IIIa, IV, VI, VIII and IX) and core proteins (V, VII, X and TP), see text. As the structure of the nucleoprotein core has not been established, the polypeptides associated with the DNA are shown in hypothetical locations.
(Adapted from Stewart, P.L. and Burnett, R.M. (1993). Jpn J. Appl. Phys., 32, 13421347). (Right) Fowl adenovirus 9 particle negatively stained with uranyl acetate, showing the characteristic double fibers of fowl adenoviruses. (From Gelderblom, H. and Maichle-Lauppe, I. (1982). Arch. Virol.72, 289298; with permission.) The bar represents 100 nm.
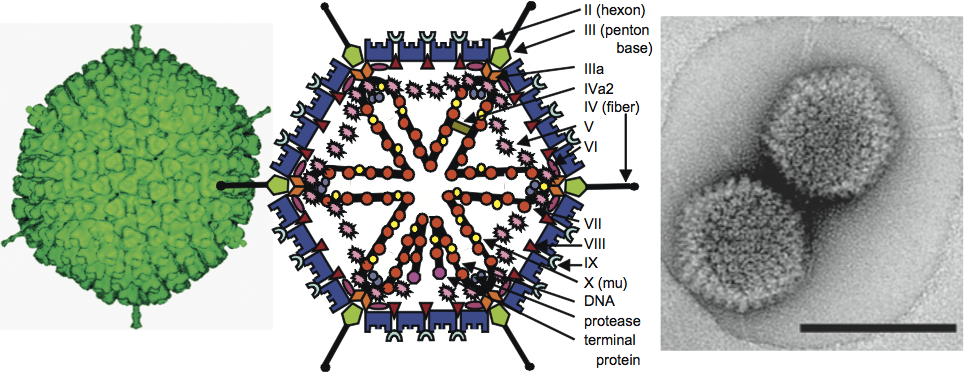
Figure 2 Schematic illustration of the various genome organizations found in members of four adenovirus genera. Black arrows depict genes conserved in every genus, grey arrows show genes present in more than one genus, and coloured arrows show genus-specific genes.
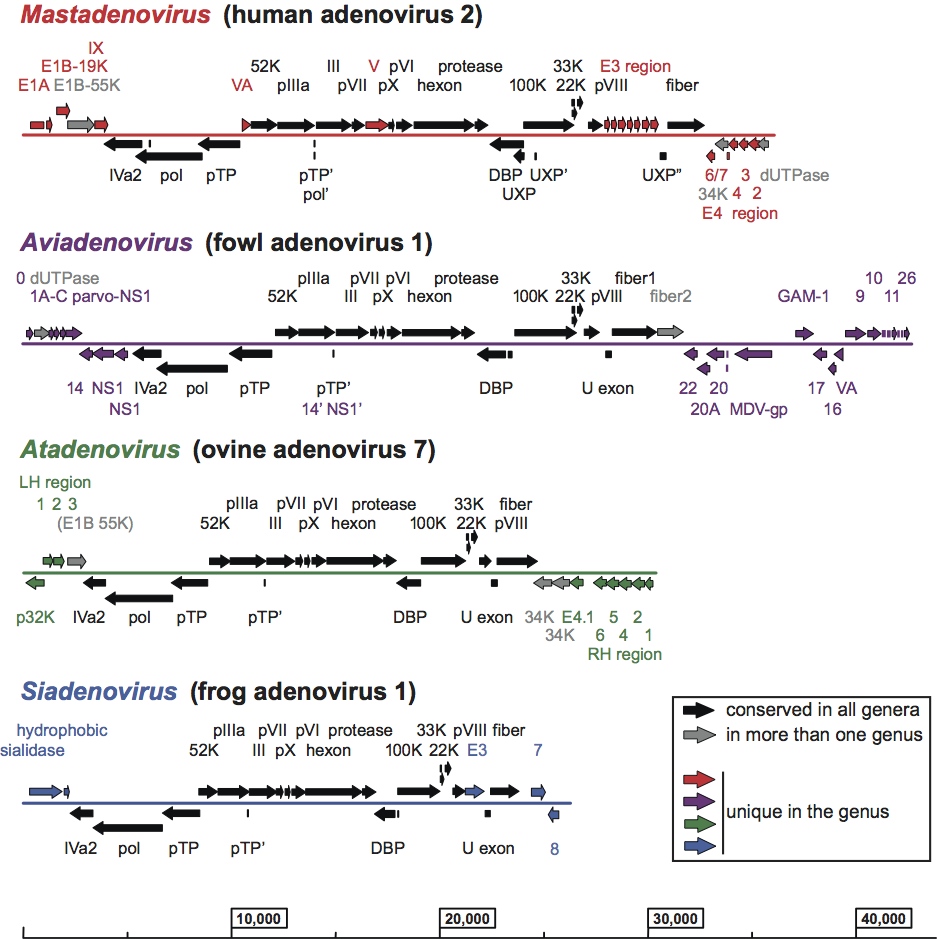
Figure 3 Schematic illustration of the transcription pattern of human adenovirus 2. The parallel lines indicate the linear dsDNA genome of 36 kbp. The dots, broken lines and split arrows indicate the spliced structures of the mRNAs. E1A, E1B, E2A, E2B, E3 and E4 refer to early transcription units. Most (but not all) late genes are in the major late transcription unit which initiates after the E1B and protein IX genes of the r strand (transcribed rightward), and which includes the L1, L2, L3, L4 and L5 families of mRNAs. Intermediate genes (of protein IX and protein IVa2) are marked with a star.
(Adapted from Wold, W.S. and Gooding, L.R. (1991). Virology, 184, 18.)
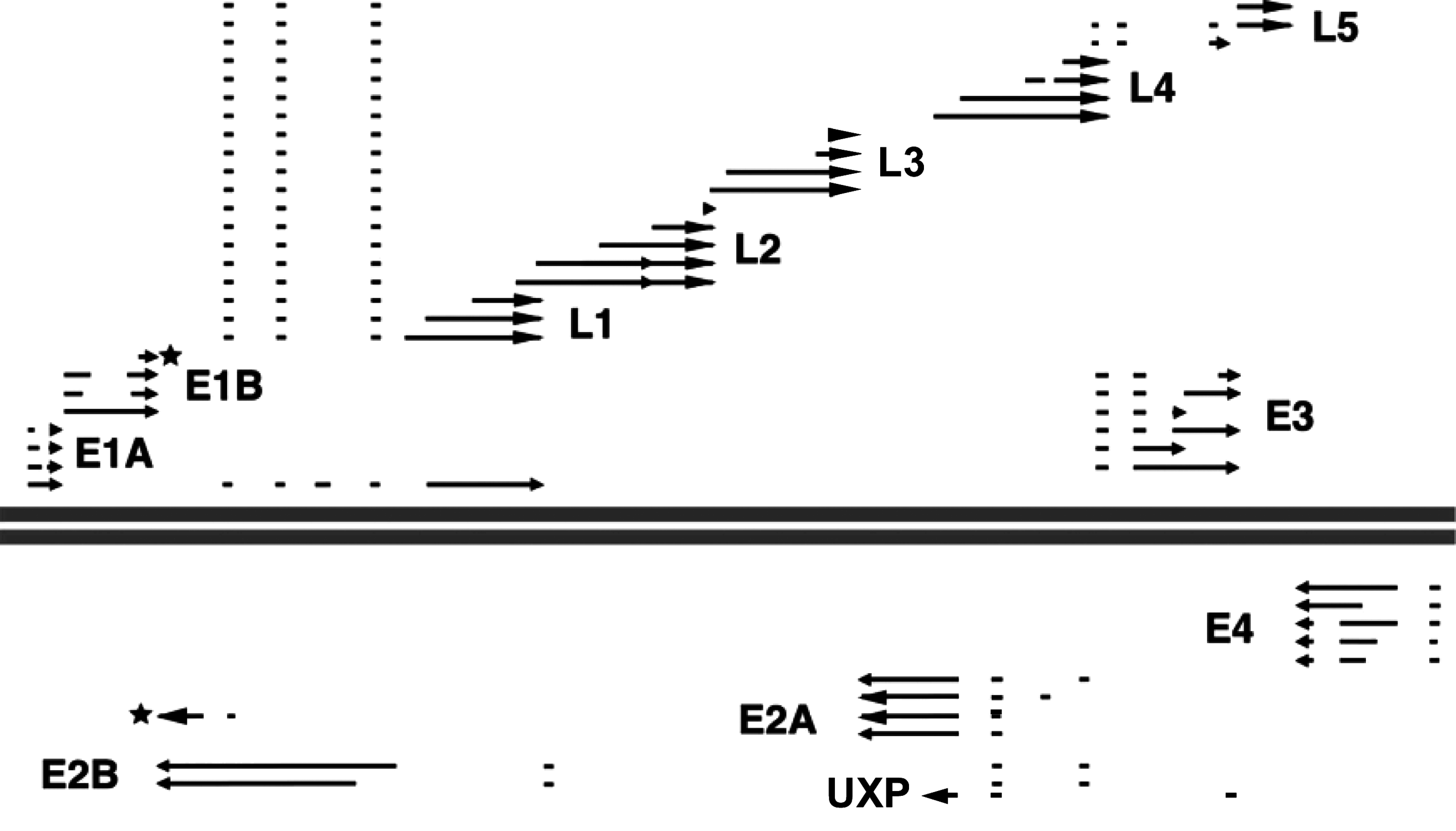
Figure 4 Cryo-EM image of ovine atadenovirus 7 at 10.6 resolution. The capsid surface is oriented around one of the three-fold axes. Penton complexes are marked in yellow, indicating the approximate bounds of the icosahedral facet. A major capsid protein (LH3) that is a key distinguishing feature of the virus is marked in blue. The bar represents 10 nm.
(Reproduced with permission from Pantelic, R.S. et al. (2008), J. Virol., 82, 73467356.)
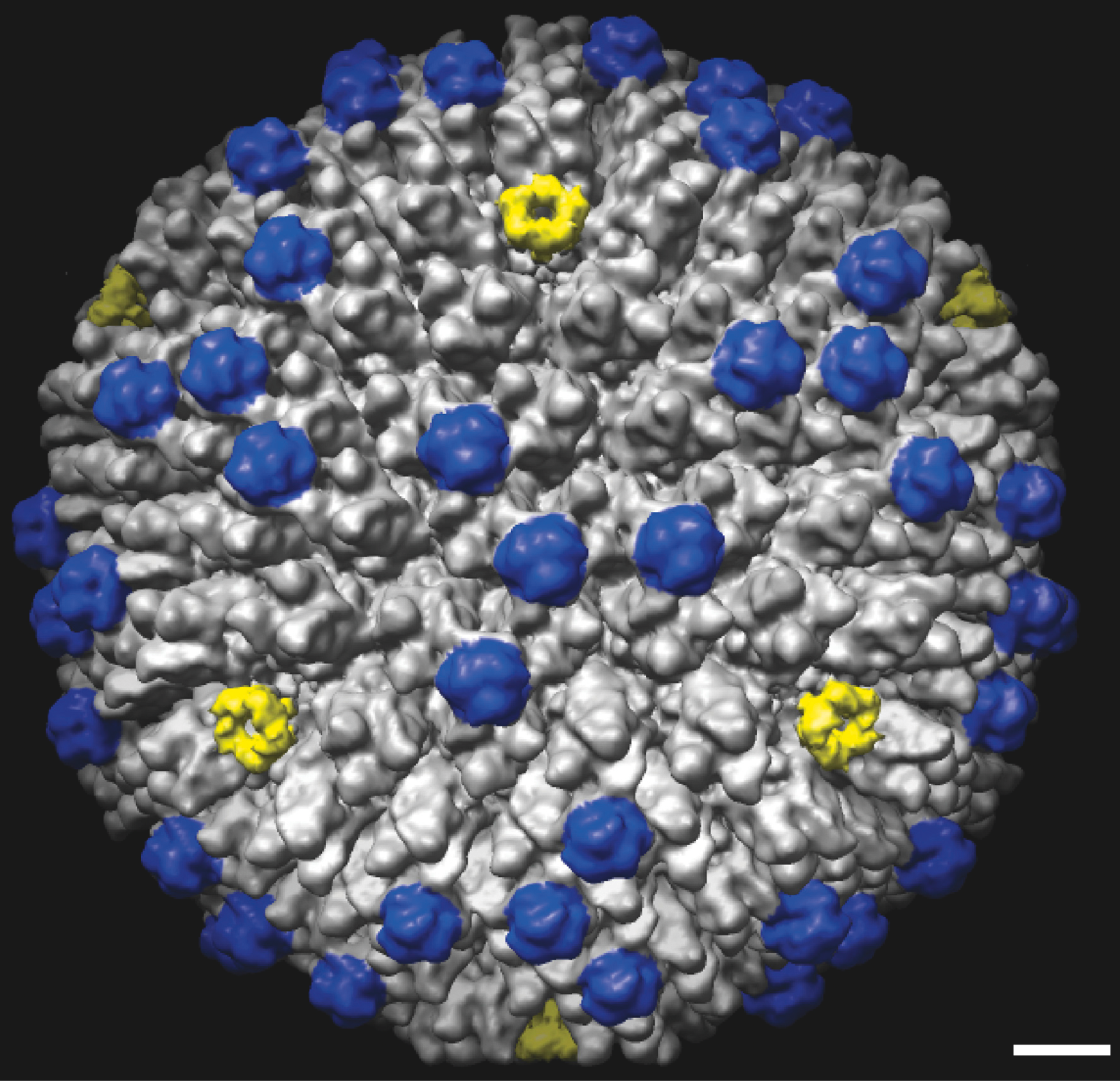
Figure 5 Phylogenetic tree of adenoviruses based on a distance matrix analysis of hexon amino acid sequences. Primate adenovirus hexons known to have resulted from homologous recombination were excluded from the analysis. Only selected members of primate adenovirus species with more than three members are shown (boxed in grey). The Seqboot (bootstrap), Protdist (categories matrix), Fitch (global rearrangements), and Consense programs of the PHYLIP 3.68 package were used. The tree was generated by unrooted calculation, and is shown with white sturgeon adenovirus 1 chosen as outgroup. Species names are indicated, but for reasons of presentation the word adenovirus is abbreviated to AdV followed by a hyphen. Abbreviations after the type numbers of simian adenoviruses: bo, bonobo; ch, chimpanzee; go, gorilla; cy, cynomolgus macaque; rh, rhesus macaque. Bootstrap values higher than 50 (from 100 re-samplings) are shown for every confirmed branching.