Family: Pseudoviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Pseudoviridae is a family of retrotransposable elements, primarily identified by genome sequencing. Pseudoviruses are often referred to as LTR-retrotransposons of the Ty1-copia family. They replicate via a virus-like intermediate referred to as a virus-like particle (VLP). VLPs do not display infectivity according to the traditional virological definition. However, there is good evidence that these particles are essential and direct intermediates in the life cycle of these elements. We will use the terms “virion” and “virus” here to conform to the usage in this volume.
Members of the family Pseudoviridae are typified by somewhat irregularly shaped VLPs that are round to ovoid, often with electron-dense centers. Although the VLPs are irregular in their native state, by expressing truncated forms of the major coat protein (Gag) icosahedral VLPs with regularity can be observed by cryo-electron microscopy (Figure 1). Saccharomyces cerevisiae Ty1 virus (Ty1) and Drosophila melanogaster copia virus (copia) both make similar looking particles, but Ty1 particles are cytoplasmic, whereas those of copia are nuclear. The typical mean radius of the VLPs is 30–40 nm. There is no envelope, although some members encode an env-like gene whose function is unknown.
Physicochemical and physical properties
In most systems, virions are only partially characterized biochemically.
Nucleic acid
The major virion RNA species consist of an LTR-to-LTR transcript of about 5–9 kb. In addition, most viruses package one or more host-derived primer tRNAs. The LTR-to-LTR transcript encodes one to two ORFs in most viruses, the equivalents of retroviral gag and pol; the second ORF is typically expressed at lower levels than the first. There is RNA, as well as various forms of DNA (intermediates) in the virion preparation. The RNA is 5–9 kb, positive sense, capped and polyadenylated; the DNA is 5.5–10 kbp. The linear ssRNA is packaged in virions, and the linear dsDNA “provirus” is integrated into the host genome.
Proteins
Both Gag and Gag-Pol primary translation products are processed by the cognate protease into final products. The known gag-encoded proteins include analogs of retroviral capsid (CA) and nucleocapsid (NC), although the latter is only present in a subset of these viruses. The known pol-encoded proteins include the protease (PR), integrase (IN), and reverse transcriptase/RNase H (RT). All of these proteins appear to be required for replication.
Lipids
None present.
Carbohydrates
None present.
Genome organization and replication
Most viruses encode a single ORF with similarity to retroviral gag and pol (Figure 2). The yeast Ty1, Ty2 and Ty4 viruses contain pol in the +1 frame relative to gag. For copia, gag is encoded on a spliced 2 kb mRNA, and differential splicing is the mechanism by which Gag and Gag-Pol stoichiometry is regulated. The mechanism(s) that regulates Gag and Gag-Pol expression for most single ORF viruses is unknown.
The virion-associated RT mediates the conversion of the LTR to LTR transcript into a full-length nucleic acid duplex containing full-length LTR sequences in the form of dsDNA. This DNA is then integrated into host DNA by the IN protein, where it becomes a part of the host genome and can persist there, essentially indefinitely. The integrated form (equivalent to the retroviral provirus) is then transcribed by host RNA polymerase II to generate new virus RNAs. In most viruses, the reverse transcription and integration processes closely mimic the replication of retroviral RNA, but there are some important exceptions.
Antigenic properties
No information available.
Biological properties
Pseudoviruses are commonly referred to as LTR retrotransposons of the Ty1/copia family. They form an intrinsic and significant part of the genome of many eukaryotic species, especially plants. For most of these viruses, the virion is an essential part of their multiplication cycle, but is not infectious, in the traditional virological sense, under normal conditions.
Species demarcation criteria in the family
In general, viruses inhabiting different host species will be considered different species, because they will have diverged through vertical descent from a common ancestor at least as much as the host species themselves (probably more so, due to the error-prone mechanism of replication). However, there are instances in the family Pseudoviridae and the family Metaviridae in which one finds two closely related viruses inhabiting the same host species, e.g. Ty1 and Ty2 of S. cerevisiae. In this pair of viruses, the RT aa sequences are quite similar. However, the capsid-encoding sequences are significantly diverged (e.g. <50% aa sequence identity; their DNA sequences fail to cross-hybridize). The question then arises whether these represent different species or more subtle variants. We have considered such viruses separate species if at least one of the major coding regions (e.g. capsid) is <50% identical to the reference aa sequence. For example, Ty1 and Ty2 Gag aa sequences are 49% identical.
Genus Pseudovirus
Type species Saccharomyces cerevisiae Ty1 virus
Virion properties
Virions are ovoid to spheroid strictly intracellular particles, sometimes observed as paracrystalline clusters in the cytoplasm in yeast cells. Particles are quite heterodisperse structurally. However, the C-terminal deletion mutant containing residues 1–381 of the Gag protein is less heterodisperse than the wild-type, and cryo-electron micrographs of these particles reveal a surface structure with icosahedral symmetry (Figure 1). The mean radius of the virions is 20–30 nm for wild type and 20 nm for the much better characterized and more uniform 1-381 mutant. The virions are not enveloped. The total Mr is estimated at 14 MDa and the sedimentation coefficient is estimated at about 200–300S for wild type and 115S for 1-381 mutant. The virions are sensitive to high temperature (65 °C).
The major virion RNA species consist of an LTR-to-LTR transcript of 5.6 kb. In addition Ty1 packages host-derived primer tRNAiMet. Ty1 particles contain two RNA molecules. This transcript encodes two overlapping ORFs, GAG and POL; the second expressed as the result of a +1 frameshift relative to the first. The major nucleic acid forms are RNA in the VLP, as well as various DNA forms (intermediates) in VLP preparations, and finally, after integration is complete, the integrated DNA “provirus”. The genome size for Ty1 is RNA – 5.6 kb; DNA – 5.9 kb. There is linear ssRNA in the virion, and the provirus (integrated) consists of dsDNA.
The known GAG-encoded proteins are the CA and a short C-terminal peptide (the latter is only inferred to exist and has not been directly observed). No recognizable NC peptide has been identified, although the latter peptide performs similar functions. The known POL-encoded proteins are a protease (PR), integrase (IN) and reverse transcriptase/RNase H (RT). All of these proteins are required for replication.
Genome organization and replication
The DNA form of the Ty1 genome consists of two 335 bp LTRs largely flanking a central coding region, although Ty1 is unusual in that GAG initiates within the U5 region of the 5’ LTR. The LTR sequences can be divided into three segments called U3, R and U5 (Figure 2). The U3 region is unique to the 3’ end of the RNA, the R region is repeated in the RNA and the U5 region is unique to the 5’ end of the RNA. Ty1 encodes a 5.6 kb ssRNA with two ORFs. The first ORF is GAG and encodes the major CA as well as a small C-terminal peptide; these two proteins are proteolytically derived from the GAG primary translation product. The second ORF, which overlaps GAG in the +1 frame, is POL and encodes protease (PR), integrase (IN), and reverse transcriptase/RNAse H (RT). These three POL proteins are derived by proteolysis of the GAG-POL precursor by the Ty1 PR; there is evidence that cleavage at the GAG-PR boundary obligatorily precedes the other cleavages. The GAG-POL precursor is expressed by an inefficient programmed frameshift that occurs within the sequence CUU AGG C (the indicated codon boundaries represent the GAG frame), which is necessary and sufficient to specify the +1 frameshift. A minor shorter transcript of about 2.2 kb is reported to be 5’ coterminal with the major transcript, but has not been fully characterized. A second minor transcript is reported to be 3’ coterminal with the major transcript and is most clearly observed in yeast strains with spt3 mutations. These spt3 mutations eliminate or greatly reduce the abundance of the full-length transcript.
The initial step in replication is transcription of the Ty1 virus to generate the full-length RNA described above. This RNA is encapsidated in the cytoplasm in a precursor particle consisting of unprocessed GAG and GAG-POL proteins. Action of the Ty1 PR then converts this into a mature virus particle. It is thought that this somehow activates the reverse transcription process.
The first step in the reverse transcription process is the extension of the tRNAiMet primer, which binds to the (−) strand primer binding site ((−)PBS) in the full-length RNA. The product of this extension is referred to as (−) strand strong stop DNA or (−)ssDNA by analogy with retroviruses. The (−)ssDNA is transferred to the 3’ end of the full-length RNA, where it can be further extended to generate a nearly full-length (−) strand DNA. Priming of the plus strand initiates at the (+) PPT1 (for polypurine tract 1) adjacent to the 3’ LTR, but the mechanism of this priming and the exact nature of the primer have not been determined. Extension yields a product, (+)ssDNA that corresponds to the similarly named retroviral intermediate. Transfer of (+)ssDNA to the left end of the (−) strand DNA sets up a primer-template that can be extended, in principle, to generate full-length duplex DNA. However, studies indicate that the (+) strand is not continuous because a second priming event occurring near the middle of the molecule at a site called (+) PPT2 results in a discontinuity in the middle of the plus strand. We refer to this final product of reverse transcription as the dsDNA form, although some experiments suggest that in many of these molecules there may be stretches of RNA rather than DNA in the (+) strand. The dsDNA is imported into the cell nucleus, possibly by a nuclear localization signal found at the C-terminus of IN.
The full length dsDNA is a substrate for the Ty1 IN, which inserts the dsDNA into a chromosomal target site, in the process generating a 5 bp target site duplication of the host target site DNA. Sequences located upstream of RNA polymerase III-transcribed genes represent strongly preferred targets for such Ty1 integration in vivo.
Biological properties
Retrotransposon Ty1 is best thought of as a genome parasite of Saccharomyces cerevisiae. In typical wild strains of this yeast, 3–15 copies of Ty1 are found per haploid genome, whereas in typical laboratory isolates, there are 25–40 copies. In addition to these complete copies, the genome contains several hundred solo-LTRs (not associated with a central coding region) or fragments thereof. Ty1 appears to be restricted to this host species and to very closely related species of Saccharomyces, but is absent from more distant species of the genus. However, those species are likely to harbor related viruses. Transmission is likely to be exclusively vertical, and horizontally through conjugation. Ty1 and/or the closely related species, Ty2, has been found in virtually all isolates of S. cerevisiae, from all over the world. No cytopathic effects have been reported. Some strains contain large numbers of Ty1 virus particles and are otherwise normal in every way. However, overexpression of Ty1 proteins leads to slow growth, but this phenotype is poorly characterized.
List of species in the genus Pseudovirus
| Arabidopsis thaliana Art1 virus |
|
|
| Arabidopsis thaliana Art1 virus | [Y08010] | (AthArt1V) |
| Arabidopsis thaliana AtRE1 virus |
|
|
| Arabidopsis thaliana AtRE1 virus | [AB021263] | (AthAtRV) |
| Arabidopsis thaliana Evelknievel virus |
|
|
| Arabidopsis thaliana Evelknievel virus | [AF039373] | (AthEveV) |
| Arabidopsis thaliana Ta1 virus |
|
|
| Arabidopsis thaliana Ta1 virus | [X13291] | (AthTa1V) |
| Brassica oleracea Melmoth virus |
|
|
| Brassica oleracea Melmoth virus | [Y12321] | (BolMelV) |
| Cajanus cajan Panzee virus |
|
|
| Cajanus cajan Panzee virus | [AJ000893] | (CcaPanV) |
| Glycine max Tgmr virus |
|
|
| Glycine max Tgmr virus | [U96748] | (GmaTgmV) |
| Hordeum vulgare BARE-1 virus |
|
|
| Hordeum vulgare BARE-1 virus | [Z17327] | (HvuBV) |
| Nicotiana tabacum Tnt1 virus |
|
|
| Nicotiana tabacum Tnt1 virus | [X13777] | (NtaTnt1V) |
| Nicotiana tabacum Tto1 virus |
|
|
| Nicotiana tabacum Tto1 virus | [D83003] | (NtaTto1V) |
| Oryza australiensis RIRE1 virus |
|
|
| Oryza australiensis RIRE1 virus | [D85597] | (OauRirV) |
| Oryza longistaminata Retrofit virus |
|
|
| Oryza longistaminata Retrofit virus | [U72726] | (OloRetV) |
| Physarum polycephalum Tp1 virus |
|
|
| Physarum polycephalum Tp1 virus | [X53558] | (PpoTp1V) |
| Saccharomyces cerevisiae Ty1 virus |
|
|
| Saccharomyces cerevisiae Ty1 virus | [M18706] | (SceTy1V) |
| Saccharomyces cerevisiae Ty2 virus |
|
|
| Saccharomyces cerevisiae Ty2 virus | [M19542] | (SceTy2V) |
| Saccharomyces cerevisiae Ty4 virus |
|
|
| Saccharomyces cerevisiae Ty4 virus | [M94164] | (SceTy4V) |
| Solanum tuberosum Tst1 virus |
|
|
| Solanum tuberosum Tst1 virus | [X52387] | (StuTst1V) |
| Triticum aestivum WIS-2 virus |
|
|
| Triticum aestivum WIS-2 virus | [CT009735] | (TaeWis1V) |
| Zea mays Hopscotch virus |
|
|
| Zea mays Hopscotch virus | [U12626] | (ZmaHopV) |
| Zea mays Sto-4 virus |
|
|
| Zea mays Sto-4 virus | [AF082133] | (ZmaStoV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Pseudovirus but have not been approved as species
None reported.
Genus Hemivirus
Type species Drosophila melanogaster copia virus
Distinguishing features
For copia, Gag is encoded on a spliced 2 kb mRNA, and differential splicing is the mechanism by which Gag and Gag-Pol stoichiometry is regulated. As in the genus Pseudovirus, these viruses encode gag and pol in one or two reading frames. The mechanism(s) that regulates Gag and Gag-Pol expression for most single ORF viruses remain to be determined.
Species in this genus typically use an initiator methionine tRNA as the primer for minus-strand DNA synthesis during reverse transcription. Hemiviruses, unlike members of the families Retroviridae and Metaviridae and genus Pseudovirus, use an initiator methionine tRNA half-molecule as a primer (or an Arg tRNA half-molecule in the case of Candida albicans Tca2 virus (Tca2)). This tRNA fragment is generated by cleaving the initiator tRNA in the anticodon stem. However, relatively little is known about the detailed mechanism of this reaction.
Both Gag and Gag-Pol primary translation products are processed by the cognate protease into final products. The known gag-encoded proteins include analogs of retroviral CA and NC. The known pol-encoded proteins include the protease (PR), integrase (IN) and reverse transcriptase/RNase H (RT). All of these proteins appear to be required for replication.
These viruses are found worldwide in fungi, algae and insects genomes. Their mode of transmission is unknown although presumed to be through vertical inheritance.
List of species in the genus Hemivirus
| Aedes aegypti Mosqcopia virus |
|
|
| Aedes aegypti Mosqcopia virus | [AF134899] | (AaeMosV) |
| Candida albicans Tca2 virus |
|
|
| Candida albicans Tca2 virus | [AF050215] | (CalTca2V) |
| Candida albicans Tca5 virus |
|
|
| Candida albicans Tca5 virus | [AF065434] | (CalTca5V) |
| Drosophila melanogaster 1731 virus |
|
|
| Drosophila melanogaster 1731 virus | [X07656] | (Dme1731V) |
| Drosophila melanogaster copia virus |
|
|
| Drosophila melanogaster copia virus | [X04456] | (DmeCopV) |
| Saccharomyces paradoxus Ty5 virus |
|
|
| Saccharomyces paradoxus Ty5 virus | [U19263] | (SceTy5V) |
| Volvox carteri Lueckenbuesser virus |
|
|
| Volvox carteri Lueckenbuesser virus | [U90320] | (VcaLeuV) |
| Volvox carteri Osser virus |
|
|
| Volvox carteri Osser virus | [X69552] | (VcaOssV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Hemivirus but have not been approved as species
None reported.
Genus Sirevirus
Type species Glycine max SIRE1 virus
Distinguishing features
All members of this genus have thus far been identified only in plants. A reverse transcriptase phylogenetic tree separates the sireviruses from pseudoviruses and hemiviruses (Figure 4). Based on the sequence of putative primer binding sites, most species in this genus likely use the acceptor stem of an initiator methionine tRNA as the primer for minus-strand DNA synthesis during reverse transcription, similar to the pseudoviruses. Clusters of conserved PPTs are also present in many species, and highly conserved DNA sequence motifs of unknown function are present in the LTRs and internal domain.
A unique ORF and multiple gene expression mechanisms characterize members of this genus. For example, viruses such as Glycine max SIRE1 virus (SIRE1), Arabidopsis thaliana Endovir virus (Endovir) and Lycopersicon esculentum ToRTL1 virus (ToRTL1) have an extra ORF between pol and the 3’ LTR (Figure 3). This ORF has conserved transmembrane domains and is referred to as an env-like ORF. For SIRE1, env-like expression is regulated by stop codon suppression. However, stop codon suppression does not seem to be utilized by the other viruses (e.g. Endovir and ToRTL1), because the env-like ORF is separated from pol by non-coding sequence. In addition, based on ORF organization, two different Gag-Pol expression mechanisms are likely utilized by members of this genus. Some viruses such as SIRE1 and Endovir encode gag and pol in a single ORF. In contrast, for viruses such as Opie-2 and Prem-2, pol resides in a +1 frame (Figure 3). A unifying characteristic of this genus is that in all members, the gag gene is nearly twice the size of gag encoded by pseudoviruses and hemiviruses.
List of species in the genus Sirevirus
| Arabidopsis thaliana Endovir virus |
|
|
| Arabidopsis thaliana Endovir virus | [AY016208] | (AthEndV) |
| Glycine max SIRE1 virus |
|
|
| Glycine max SIRE1 virus | [AF053008] | (GmaSIRV) |
| Lycopersicon esculentum ToRTL1 virus |
|
|
| Lycopersicon esculentum ToRTL1 virus | [U68072] | (LesToRV) |
| Nicotiana debneyi Tnd1 virus |
|
|
| Nicotiana debneyi Tnd1 virus | [AF059674] | (NdeTnd1V) |
| Zea mays Opie-2 virus |
|
|
| Zea mays Opie-2 virus | [U68408] | (ZmaOp2V) |
| Zea mays Prem-2 virus |
|
|
| Zea mays Prem-2 virus | [U41000] | (ZmaPr2V) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Sirevirus but have not been approved as species
| Oryza sativa Osr7 virus | [AP002538] | (OsaOsr7V) |
| Oryza sativa Osr8 virus | [AC021891] | (OsaOsr8V) |
List of unassigned species in the family Pseudoviridae
| Phaseolus vulgaris Tpv2-6 virus |
|
|
| Phaseolus vulgaris Tpv2-6 virus | [AJ005762] | (PvuTpvV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
Phylogenetic relationships within the family
Phylogenetic analysis of the reverse transcriptase domain (Figure 4) does not support the current taxonomy, although the sireviruses clearly form a natural grouping.
Similarity with other taxa
Like the families Hepadnaviridae and Metaviridae, the Pseudoviridae are clearly related to the family Retroviridae. All four families are linked by reverse transcription and a viral core structure made up of Gag-like proteins. Pseudoviruses are different from the other two families in that they have an unusual organization (PR-IN-RT-RH) of the gene pol. Members of the families Pseudoviridae, Metaviridae and Retroviridae also share the following: a proviral form characterized by LTRs: protease, reverse transcriptase, RNase H and integrase activities essential for multiplication; readthrough-mediated gag-pol gene expression (in some species); and tRNA primers (in most species).
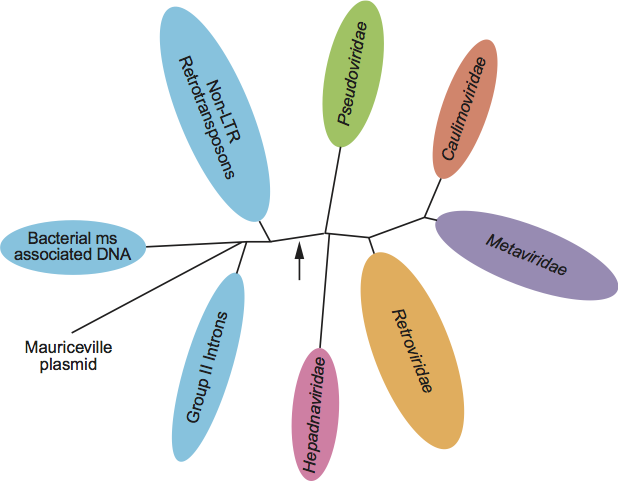
An important and controversial question is the extent of the relationship between the families Pseudoviridae, Metaviridae and the Retroviridae. Because the genomic structures of viruses in families Pseudoviridae and Metaviridae are clearly related to, but typically simpler than the viruses in the family Retroviridae, many authors who have considered the problem have concluded that the families Pseudoviridae and Metaviridae represent more primitive groups; the family Metaviridae probably spawned the members of the family Retroviridae (presumably by incorporating genes encoding ligands for cell-surface receptors). This conclusion makes sense within the context of the enormous diversity of other types of retroelements, which are all clearly phylogenetically related by the presence of RT (Figure 5), but not all of which encode a virus-like intermediate. An alternative viewpoint that cannot be ruled out, but for which there is less support, is that members of the family Metaviridae represent degenerate forms of the family Retroviridae.
Derivation of names
Hemi: from Greek hemi, “half”, referring to the half-molecule of tRNA used as a primer for reverse transcription.
Pseudo: from Greek pseudo, “false”, to connote some uncertainty as to whether these are true viruses.
Sire: from the abbreviation of the species name: Glycine max SIRE1 virus (SIRE).
Further reading
Boeke, J.D. and Sandmeyer, S.B. (1991). Yeast transposable elements. In: Broach, J., Jones, E. and Pringle, J. (Eds.), The Molecular and Cellular Biology of the Yeast Saccharomyces, vol. 1. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, pp. 193-261.
Boeke, J.D. and Stoye, J.P. (1996). Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Varmus, H., Hughes, S. and Coffin, J. (Eds.), Retroviruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, pp. 343-436.
Garfinkel, D.J., Boeke, J.D. and Fink, G.R. (1985). Ty element transposition: reverse transcriptase and virus-like particles. Cell, 42, 507-517.
Kikuchi, Y., Ando, Y. and Shiba, T. (1986). Unusual priming mechanism of RNA-directed DNA synthesis in copia retrovirus-like particles of Drosophila. Nature, 323, 824-826.
Kumar, A. and Bennetzen, J.L. (2000). Plant retrotransposons. Annu. Rev. Genet., 33, 479-532.
Laten, H.M., Majumdar, A. and Gaucher, E.A. (1998). SIRE-1, a copia/Ty1-like retroelement from soybean, encodes a retroviral envelope-like protein. Proc. Natl Acad. Sci., U S A, 95, 6897-902.
Mellor, J., Malim, M.H., Gull, K., Tuite, M.F., McCready, S., Dibbayawan, T., Kingsman, S.M. and Kingsman, A.J. (1985). Reverse transcriptase activity and Ty RNA are associated with virus-like particles in yeast. Nature, 318, 583-586.
Palmer, K.Y., Tichelaar, W., Myers, N., Burns, N.R., Butcher, S.J., Kingsman, A.J., Fuller, S.D. and Saibil, H.R. (1997). Cryo-EM structure of yeast Ty retrotransposon virus-like particles. J. Virol., 71, 6863-6868.
Peterson-Burch, B.D. and Voytas, D.F. (2002). Genes of the Pseudoviridae (Ty1/copia Retrotransposons). Mol. Biol. Evol., 19, 1832-1845.
Voytas, D.F. and Boeke, J.D. (2002). Ty1 and Ty5. In: Craig, N. (Ed.), Mobile DNA II. American Society for Microbiology, Washington, DC, pp. 631-662.
Xiong, Y. and Eickbush, T.H. (1990). Origin and evolution of retroelements based on their reverse transcriptase sequences. EMBO J., 9, 3353-3362.
Contributed by
Boeke, J.D., Eickbush, T., Sandmeyer, S.B. and Voytas, D.F.
Figures
Figure 1 Saccharomyces cerevisiae Ty1 virus (SceTy1V) 1-381 virions; surface structure of two forms (T = 3, left; T = 4, right) determined by cryo-electron microscopy, with the corresponding diagrammatic models.
(Courtesy of H. Saibil, adapted from J. Virol., 71, 68636868.)
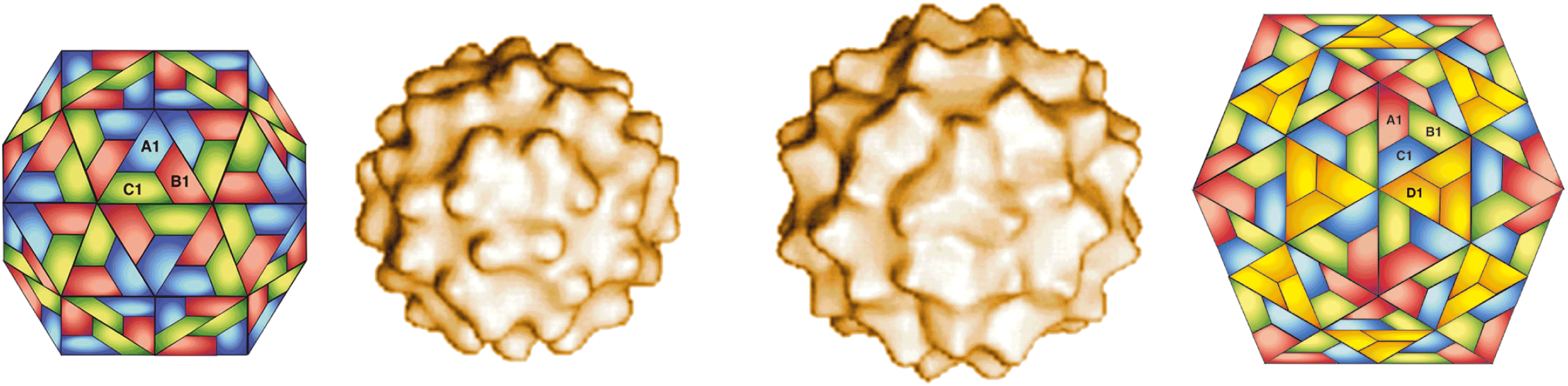
Figure 2 The genomic organization of Saccharomyces cerevisiae Ty1 virus (SceTy1V) (5.9 kb) and Drosophila melanogaster copia virus (DmeCopV) (5.1 kb). Black boxes within the LTRs depict sequences repeated at the 5 and 3 ends of the viruss transcripts (R regions); sequences 5 of R represent U3, and sequences 3 of R represent U5. Open boxes below the viruses indicate gag and pol. Conserved aa sequences in pol that identify protease (PR), integrase (IN) and reverse transcriptase/RNAse H (RT) are labeled. Individual mRNAs are depicted as arrows. Arrowhead indicates site of ribosomal frameshifting.
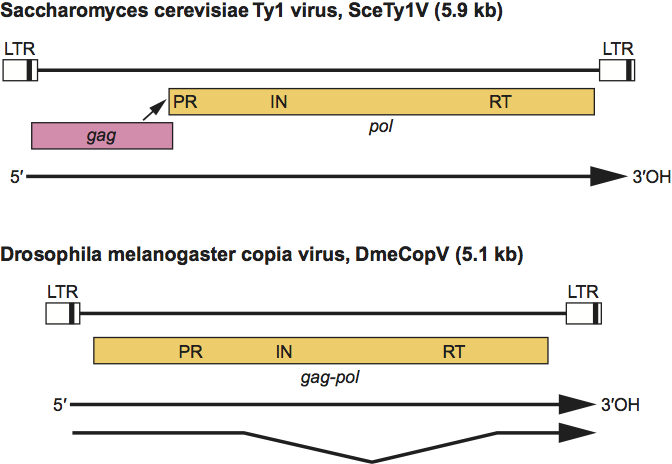
Figure 3 Examples of variability in the genome organization of members of the genus Sirevirus. Variability can be observed by the presence or absence of an env-like ORF, the organization of the env-like ORF relative to pol, and the organization of gag and pol.
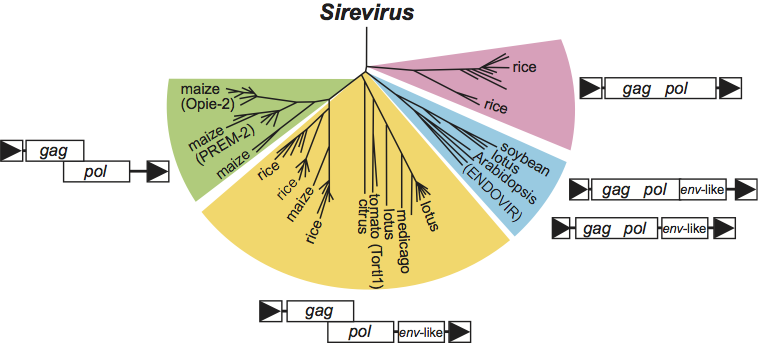
Figure 4 Phylogenetic tree based on the amino acid sequences of the reverse transcriptase domain of members of the family Pseudoviridae. The Pfam HMM reverse transcriptase model for Pseudoviridae retrotransposons (RVT_2 PF07727.6) was used to scan and locate the respective domain in each element. After the alignment, MEGA4 was used to construct the phylogenetic tree using the neighbor-joining distance method. Bootstrap support was calculated based on 1000 replicates (values shown where >50%) and the evolutionary distances were computed using the Poisson correction model. Members of the genus Pseudovirus are shown in red, Hemivirus in blue and Sirevirus in green. Tpv2-6 is an unassigned species and Osr7 and Osr8 have not yet been classified but clearly belong with the sireviruses.
(Courtesy of A. Bousios, Institute of Agrobiotechnology, Centre for Research and Technology Hellas, Thessaloniki, 57001, Greece.)
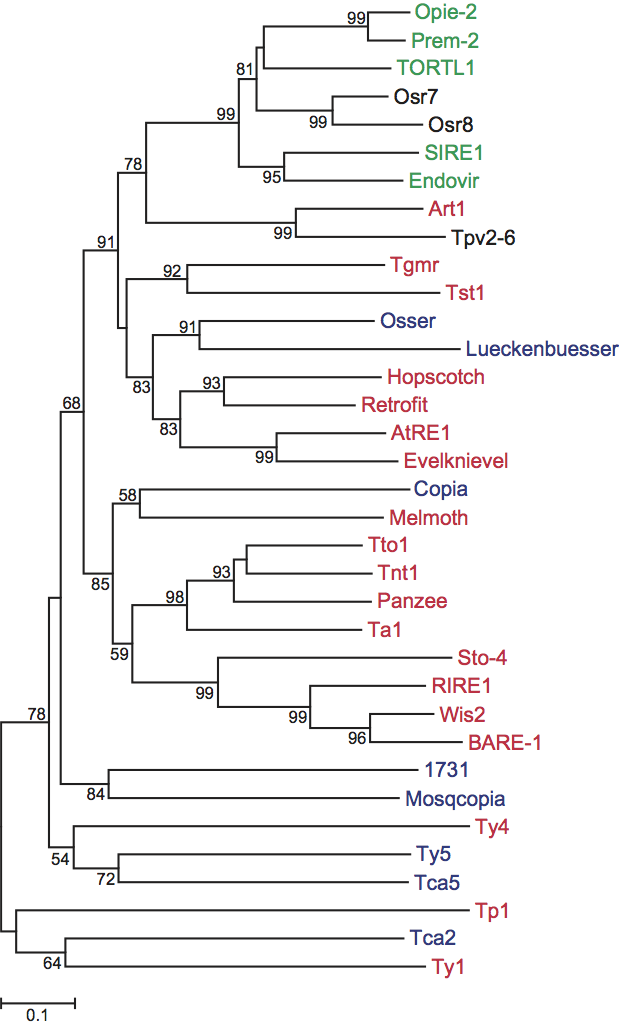
Figure 5 Unrooted phylogenetic tree of all class of reverse transcriptase containing viruses. While over 100 reverse transcriptase sequence were used to generate this phylogeny, to simplify visual comparison of the major topologies of the tree, viruses from the same class that are located on the same branch of the tree are indicated by an oval. The length of the oval corresponds to the most divergent viruses within that oval. The arrow indicates a possible root of the tree using RNA polymerase sequences.