Family: Metaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Metaviridae is a family of retrotransposons that have been found in all studied lineages of eukaryotes. These viruses, primarily identified by their ability to induce mutations or by genome sequencing, replicate via virus-like intermediates referred to as a virus-like particles (VLPs). Members of the Metaviridae family are often referred to as LTR-retrotransposons of the Ty3-gypsy family. While there is good evidence that these particles are essential and direct intermediates in the life cycle of these viruses, only in one case, Drosophila melanogaster Gypsy virus (DmeGypV), do VLPs display infectivity according to the traditional virological definition. Viruses that generate VLPs or virions will be referred to collectively in this chapter as retrotransposons.
Morphology of particles is relatively poorly characterized and capsomeric symmetry is unknown. Members include species that produce primarily or exclusively intracellular particles [e.g., Saccharomyces cerevisiae Ty3 virus (SceTy3V)] so that collections of particles are heterogeneous with respect to stage of maturation. These intracellular particles will be referred to as virus-like particles (VLPs). Extracellular particles are enveloped with ovoid cores and will be referred to as virions (e.g., DmeGypV).
Physicochemical and physical properties
In most systems virions are only crudely characterized biochemically.
Nucleic acid
The genomes of retrotransposons in this family are positive strand RNAs. The genomic RNA is polyadenylated at the 3′ end; a cap structure has not yet been described, but may be presumed, given similarities of member elements with retroviruses. In addition to the RNA genome, some cellular RNAs may be randomly associated with particles including specific tRNAs in the case of virus replication which is primed by tRNAs. Particle fractions from cells are heterogeneous with respect to maturation and so are associated with intermediates and products of reverse transcription in addition to genomic RNA.
Proteins
Proteins present in characterized VLPs include a major structural protein or capsid (CA), an aspartate protease (PR), reverse transcriptase containing an RNase H domain (RT-RH), and integrase (IN). For most viruses, these proteins are not yet characterized, but are predicted based on similarity of the protein sequence with those of proteins encoded by the internal domain. In most cases, the CA is not markedly similar to retroviral CA. VLPs of some retrotransposons in this family contain proteins with the metal finger characteristic of nucleocapsid (NC) of retroviruses. All viruses of the genus Errantivirus are distinguished by the presence of processed envelope proteins that apparently correspond to retroviral transmembrane (TM) and surface (SU) proteins. Only a limited number of viruses of the genera Metavirus and Semotivirus contain a putative envelope gene (env).
Lipids
In the case of members that generate virions, the virion membrane appears to be derived from the membrane of the host cell.
Carbohydrates
Carbohydrates have not been characterized, although their presence is inferred from sensitivity of the DmeGypV envelope precursor protein to digestion with endoglycosidase F.
Genome organization and replication
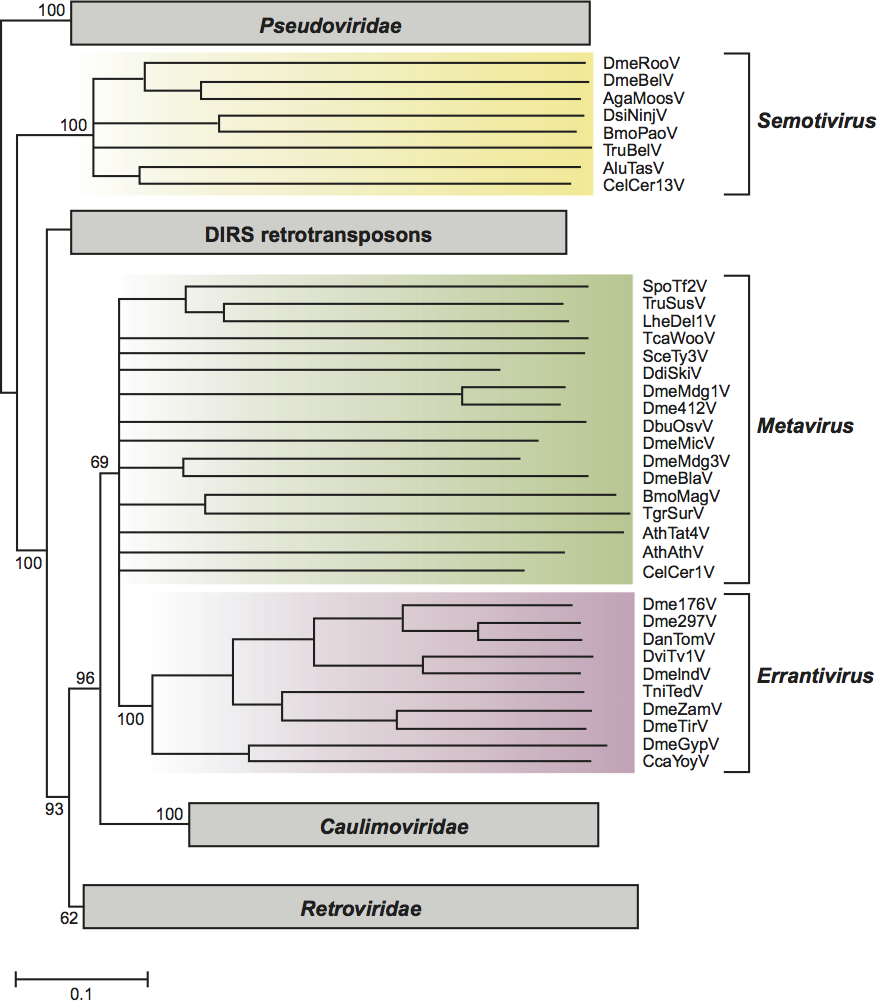
The integrated form of these retrotransposons is composed of long terminal repeats (LTRs) flanking a central unique domain (Figure 1). The length of the viral genomes ranges from 4 kbp to more than 10 kbp. The LTRs are from 77 nt in the case of the Bombyx mori mag virus (BmoMagV) to greater than 2 kbp in length, in the cases of Drosophila virilis Ulysses virus (DviUlyV) and the Tribolium castaneum Woot virus (TcaWooV). Chromosomal copies of the viruses are flanked by short direct repeats of sequences derived from the insertion site. The length of the repeat is characteristic of the virus and ranges from 4 to 6 bp. The internal domain contains one to three ORFs. The 3′ end of the final ORF can extend into the downstream LTR. In all cases, the order of domains encoded in the ORFs is inferred to be: 5′-CA-(NC where present)-PR-RT-RH-IN-3′. Where characterized, envelope proteins are encoded downstream of the IN domain by spliced mRNAs. These ORFs are referred to differently for different viruses and in this discussion, will be generally referred to as gag, pol and env. Thus viruses may have one gag-pol ORF, two (gag and pol) or three (gag, pol and env) ORFs.
Transcription of the genomic RNA is initiated in the upstream LTR and terminates at a position downstream of that site in the downstream LTR. This divides the long terminal repeats into regions represented uniquely in the 5′ end of the genomic RNA (U5), uniquely in the 3′ end of the genomic RNA (U3) or repeated at the 5′ and 3′ ends (R). Thus, the LTRs are comprised of U3-R-U5 regions analogous to those found in integrated retroviruses. By analogy with retroviruses, these species may carry two copies of the RNA genome per virion or particle; however, this has not yet been demonstrated, and dimerization functions have not yet been characterized.
Genomic RNA is translated into proteins required for particle formation, polyprotein maturation, reverse transcription and integration. Intracellular particle preparations show that particle fractions are comprised predominantly of species derived from the upstream portion of the ORF or where two or three ORFs are present from the first ORF. Where two ORFs occur, they usually overlap, and the second ORF is translated as a fusion protein of the first and second ORF translation products. The mechanism of frameshifting is not uniform among the member viruses. In case of the Schizosaccaromyces pombe Tf1 virus (SpoTf1V), the most completely characterized virus of this family containing one ORF, it appears that a polyprotein is produced and that later proteolytic events are responsible for a high ratio of major structural proteins to catalytic proteins. Little is known about where in the cell particle assembly occurs. PR is required for maturation of viral proteins. Catalytic proteins are PR, RT-RH and IN. Shortly after production of protein precursors, processed species are observed. Based on similarity of these metaviruses to retroviruses, it is likely that processing follows, and is dependent upon, intracellular assembly. Particle fractions are associated with genomic RNA and extrachromosomal DNA. RT activity associated with the particle fraction can be measured by exogenous assays.
Reverse transcription of genomic RNA of known members of this family is primed from either the 5′ end of the genomic RNA or from the 3′ end of a tRNA. In each case, the complementarity is overlapping, adjacent to, or just downstream of the U5 region of the genomic transcript. In cases in which the reverse transcription intermediates have been characterized (DmeGypV, Saccharomyces cerevisiae Ty3 virus (SceTy3V), and Schizosaccharomyces pombe Tf1 virus (SpoTf1V)), data are consistent with a species representing a minus-strand copy templated from the site of priming up to the 5′ end of the genomic RNA. This is a minor species. By analogy with retroviruses, this intermediate is probably transferred to the 3′ end of the genomic RNA, where an overlap of the R region minus strand represented in the cDNA, and the R region plus strand, represented at the 3′ end of the genomic RNA, allow transfer of the minus-strand strong stop which then acts to prime copying of the template plus-strand genomic RNA. Plus-strand priming probably occurs, as in retroviruses, from a polypurine tract or related sequence overlapping, adjacent to, or just upstream of the U3 region in the genomic RNA. This is consistent with priming from a site of cleavage by RH. Plus-strand, strong-stop species have been identified for some representatives (DmeGypV and SceTy3V) which are consistent with this position of priming and copying through to the first modified base in the primer tRNA. This family is heterogeneous with respect to the presence of extra terminal nt in the extrachromosomal replicated DNA and with respect to the presence of TG-CA inverted repeats at the ends of the integrated sequence.
Biological properties
Activation of transposition of these viruses can cause disruption of host physiology depending on the site of insertion. Several members exhibit preferential patterns of insertion. It is notable that germline activation, which is a feature of some retroviruses, also occurs for some of these viruses. For example, SceTy3V transcription is induced by mating pheromone and transposition occurs after mating. In the case of the DmeGypV and the Drosophila melanogaster Zam virus (DmeZamV), transposition occurs in germline cells.
Genus Metavirus
Type species Saccharomyces cerevisiae Ty3 virus
Distinguishing features
Saccharomyces cerevisiae Ty3 virus (SceTy3V) forms generally spherical, but irregular, intracellular particles of about 50 nm in diameter. These are observed as clusters or as individual particles in the cytoplasm of cells expressing high levels of SceTy3V RNA. Particles sediment as a heterodisperse population around 156S. The major particle-associated RNA species is a 5.2 kb, polyadenylated RNA. The primer of minus-strand reverse transcription is tRNAiMet, which is complementary to a primer binding site (pbs). A minor 3.1 kb species is also observed, but the extent to which this is associated with particles has not been characterized. The 5.4 kb RNA contains two ORFs analogous to gag and pol, GAG3 and POL3, which overlap in the +1 frame.
The genomic RNA is translated into Gag3 and Gag3-Pol3 polyproteins which are processed by SceTy3V PR. Mature proteins include CA (26 kDa), NC (15 kDa), PR (15 kDa), RT-RH and IN (58 kDa and 61 kDa, respectively), and an RT-RH-IN fusion protein of approximately 115 kDa. The molecular composition of the RT is not yet known. A protein of 10 kDa is predicted to be encoded between PR and RT but has not been identified. The integrated form of SceTy3V is 5.4 kbp in length and consists of an internal domain flanked by two LTRs (sigma elements) 340 bp in length. Insertions of SceTy3V are flanked by 5 bp direct repeats derived from insertion site cleavage and repair. SceTy3V is transcribed into a 5.2 kb genomic RNA. The tRNAiMet pbs has its 5′ end two nt downstream of the junction of the 5′ LTR with the internal domain. The full-length SceTy3V DNA molecule is 2 bp longer at each end than the integrated molecule, consistent with predictions based on the positions of the priming sequences. Two nt are removed from each 3′ end prior to integration. SceTy3V integrates within one or two nt of the transcription initiation site of genes transcribed by RNA polymerase III. SceTy3V is found in one to five copies in typical laboratory strains of Saccharomyces cerevisiae. In addition, there are approximately 30-40 copies of the isolated LTRs present. The latter presumably arose by recombination between the LTRs of complete viruses. SceTy3V transcription is induced by pheromone signal transduction. Although proteins are produced in cells undergoing signaling, DNA is not made in cells arrested in G1 of the cell cycle. Consequently, it is most likely that in natural populations, transposition only occurs after the fusion of mating cells to form diploids.
Species demarcation criteria in the genus
Although the members of this family encode Pol proteins which are similar to those of SceTy3V, other properties of the members are distinct from SceTy3V. Several of the elements (Drosophila melanogaster micropia virus, DmeMicV; Lilium henryi del1 virus [LheDel1V]; and Schizosaccharomyces pombe Tf1 virus [SpoTf1V], Schizosaccharomyces pombe Tf2 virus [SpoTf2V] and Cladosporium fulvum T1 virus [CfuT1V]) encode a single long ORF from which major structural proteins, as well as Pol proteins, are expressed. In the most completely characterized case, SpoTf1V, the ORF is expressed as a single polyprotein, which is processed by a mechanism dependent on the SpoTf1V PR. Cells in stationary phase have the highest ratio of major structural to catalytic protein and products of reverse transcription accumulate concomitant with this transition. Members of this genus are also distinguished from SceTy3V by aspects of replication priming. SpoTf1V forms an RNA structure involving 89 bases at the 5′ end of the RNA, which is processed by RNase H to cleave within the structure between nt 11 and 12 from the 5′ end. This cleavage allows priming of SpoTf1V RT from the 3′ end of the 11 nt fragment annealed immediately downstream of the 5′-LTR. Other members of the genus (SpoTf2V and CfuT1V) have sequences consistent with a similar mechanism of self-priming. Thus these viruses are distinguished by self-priming, i.e., by an apparent lack of requirement for 3′-end processing. Individual species in the genus all have less than 50% identity in their Gag protein sequences compared to all other species. For example, although Drosophila melanogaster Mdg virus (DmeMdg1V) and Drosophila melanogaster 412 virus (Dme412V) each infects Drosophila melanogaster, their gag sequences are only 39% identical. Two viruses, Drosophila buzzatti Osvaldo virus (DbuOsvaV) and Arabidopsis thaliana Athila virus (AthAthV), have an env-like gene. Based on this property it is possible to place these viruses within the Errantivirus genus. However, the envelope-like proteins of DbuOsvV and AthAthV are unrelated in sequence to that of the errantiviruses, and based on the sequence of their RT domain, cannot be placed within the Errantivirus genus.
List of species in the genus Metavirus
| Arabidopsis thaliana Athila virus |
|
|
| Arabidopsis thaliana Athila virus - At | [AC007209] | (AthAthV-At) |
| Arabidopsis thaliana Tat4 virus |
|
|
| Arabidopsis thaliana Tat4 - At | [AB005247] | (AthTat4V-At) |
| Bombyx mori Mag virus |
|
|
| Bombyx mori Mag virus - 200 X 300 | [X17219] | (BmoMagV-200X300 ) |
| Caenorhabditis elegans Cer1 virus |
|
|
| Caenorhabditis elegans Cer1 virus - Ce | [U15406] | (CelCer1V-Ce) |
| Cladosporium fulvum T- 1 virus |
|
|
| Cladosporium fulvum T- 1 virus - cf | [Z11866] | (CfuT1V-cf) |
| Dictyostelium discoideum Skipper virus |
|
|
| Dictyostelium discoideum Skipper virus - AX2 AX3 AX3 AX2 | [AF049230] | (DdiSkiV-AX2/3) |
| Drosophila buzzatii Osvaldo virus |
|
|
| Drosophila buzzatii Osvaldo virus - BU 30/4 | [AJ133521] | (DbuOsvV-BU30/4 ) |
| Drosophila melanogaster Blastopia virus |
|
|
| Drosophila melanogaster Blastopia virus - dm | [Z27119] | (DmeBlaV-dm) |
| Drosophila melanogaster Mdg1 virus |
|
|
| Drosophila melanogaster Mdg1 virus - dm | [X59545] | (DmeMdg1V-dm) |
| Drosophila melanogaster Mdg3 virus |
|
|
| Drosophila melanogaster Mdg3 virus - dm | [X95908] | (DmeMdg3V-dm) |
| Drosophila melanogaster Micropia virus |
|
|
| Drosophila melanogaster Micropia virus - CantonS | [X14037] | (DmeMicV-CantonS ) |
| Drosophila melanogaster 412 virus |
|
|
| Drosophila melanogaster 412 virus - dm | [X04132] | (Dme412V-dm) |
| Drosophila virilis Ulysses virus |
|
|
| Drosophila virilis Ulysses virus - dv | [X56645] | (DviUlyV-dv) |
| Fusarium oxysporum Skippy virus |
|
|
| Fusarium oxysporum Skippy virus - f. sp. lycopersici 42-87 | [L34658] | (FoxSkiV-42-87 ) |
| Lilium henryi Del1 virus |
|
|
| Lilium henryi Del1 virus - lh | [X13886] | (LheDel1V-lh) |
| Saccharomyces cerevisia Ty3 virus |
|
|
| Saccharomyces cerevisia Ty3 virus - AB950 | [M34549] | (SceTy3V-AB950 ) |
| Schizosaccharomyces pombe Tf1 virus |
|
|
| Schizosaccharomyces pombe Tf1 virus - NCYC 132 | [M38526] | (SpoTf1V-NCYC 132 ) |
| Schizosaccharomyces pombe Tf2 virus |
|
|
| Schizosaccharomyces pombe Tf2 virus - 972 | [L10324] | (SpoTf2V-972) |
| Takifugu rubripes Sushi virus |
|
|
| Takifugu rubripes Sushi virus - Tr | [AF030881] | (TruSusV-Tr) |
| Tribolium castaneum Woot virus |
|
|
| Tribolium castaneum Woot virus - A4/Ey | [U09586] | (TcaWooV-A4/Ey ) |
| Tripneustis gratilla SURL virus |
|
|
| Tripneustis gratilla SURL virus - Tg | [M75723] | (TgrSurV-Tg) |
Species names are in italic script; names of isolates and synonyms are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Metavirus but have not been approved as species
None reported.
Genus Errantivirus
Type species Drosophila melanogaster Gypsy virus
Distinguishing features
All elements contain a similar env-like ORF.
Virion properties
Expression of DmeGypV results in production of enveloped irregular particles of approximately 100 nm in diameter and also much smaller non-enveloped particles. The env gene predicts a protein of 54 kDa. The actual protein has an apparent size of 66 kDa and is N-glycosylated as indicated by susceptibility to endoglycosidase F. More rapidly migrating molecules of 54 kDa and 28 kDa are also observed and are inferred to result from proteolytic processing by host enzymes, as is the case of members of the family Retroviridae.
Genome organization and replication
DmeGypV is 7469 bp in length including two LTRs of 482 bp (Figure 1). It differs from most retroviruses and retrotransposons in that the termini are composed of AG…TT rather than TG…CA. The 11 nt immediately adjacent to the upstream U3 element and overlapping by one nt is complementary to tRNALys. The DNA flanking the insertion includes 4 bp repeats, and the insertion site preference is for YRYRYR (where Y=purine and R=pyrimidine) sequence. The genomic RNA contains one ORF encoding the major structural protein and a second ORF overlapping in the −1 frame, encoding homologs of retroviral PR, RT-RH and IN. A third ORF, env, encoding a 54 kDa envelope protein, occurs in a spliced 2.1 kb mRNA. This protein is apparently N-glycosylated, processed into smaller species, and is analogous to retroviral envelope proteins by virtue of hydrophobic putative membrane spanning domains, localization to the viral membrane, similarity of processing sites for cleavage into trans-membrane and surface domains, and glycosylation. An envelope protein of similar sequence to that in DmeGypV has been identified for all members of this genus. The Env protein of DmeZamV is translated from a 1.7 kb spliced message. The envelope proteins of errantiviruses have been shown to have sequence similarity to the viral env gene of certain baculoviruses.
DmeGypV is transcribed into a 6.5 kb genomic RNA. A minus-strand strong stop species of approximately 242 nt (with RNA removed) has been identified. Plus-strand strong-stop DNA species of 479 nt, which are similar in length to the LTR, and a species longer by 15 to 18 nt, presumed to result from copying of the tRNA primer, have been observed.
Biological properties
DmeGypV transposition is repressed by the activity of the flamenco gene. In females homozygous for the permissive allele of flam, the somatic follicle cells surrounding maternal germline cells appear to accumulate DmeGypV RNA and envelope protein. Transposition, however, is observed in the maternal germ cells, and this has led to the hypothesis that transposition is attributable to infection from surrounding follicle cells. A similar path of activity has also been suggested for DmeZamV and DmeIdeV. Infection has been demonstrated to result when susceptible strains are raised in the presence of DmeGypV particles mixed into their food. Incubation with antibodies against the Env protein decreased the level of infection, implicating Env in this process.
Species demarcation criteria in the genus
At least one member of this group, DmeGypV, is infectious, and all members of the genus Errantivirus have a similar third ORF resembling env. While this property makes the errantiviruses candidates for inclusion into the family Retroviridae rather than the family Metaviridae, there is no sequence similarity between the env genes of these two groups, suggesting independent acquisition events. In addition, examination of phylogenetic relationship of these viruses based on their reverse transcriptase sequences (Figure 2) places the errantiviruses as an independent lineage distinct from the family Retroviridae. Individual species in the genus all have less than 50% identity in their Gag protein sequences compared to all other species. For example DmeZamV and DmeGypV are two species of viruses in this family from Drosophila melanogaster, but their gag sequences are only 35% identical.
List of species in the genus Errantivirus
| Ceratitis capitata Yoyo virus |
|
|
| Ceratitis capitata Yoyo virus - Med+ | [U60529] | (CcaYoyV-Med+) |
| Drosophila ananassae Tom virus |
|
|
| Drosophila ananassae Tom virus - da | [Z24451] | (DanTomV-da) |
| Drosophila melanogaster Gypsy virus |
|
|
| Drosophila melanogaster Gypsy virus - gy | [M12927] | (DmeGypV-gy) |
| Drosophila melanogaster Idefix virus |
|
|
| Drosophila melanogaster Idefix virus - dmi | [AJ009736] | (DmeIdeV-dmi) |
| Drosophila melanogaster Tirant virus |
|
|
| Drosophila melanogaster Tirant virus - dm | [X93507] | (DmeTirV-dm) |
| Drosophila melanogaster Zam virus |
|
|
| Drosophila melanogaster virus - dm | [AJ000387] | (DmeZamV-dm) |
| Drosophila melanogaster 17.6 virus |
|
|
| Drosophila melanogaster 17.6 virus - dm | [X01472] | (Dme176V-dm) |
| Drosophila melanogaster 297 virus |
|
|
| Drosophila melanogaster 297 virus - dm | [X03431] | (Dme297V-dm) |
| Drosophila virilis Tv1 virus |
|
|
| Drosophila virilis Tv1 virus - dv | [AF056940] | (DviTv1V-dv) |
| Trichoplusia ni TED virus |
|
|
| Trichoplusia ni TED virus - mutant FP- D | [M32662] | (TniTedV-FP-D) |
Species names are in italic script; names of isolates and synonyms are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Errantivirus but have not been approved as species
None reported.
Genus Semotivirus
Type species Ascaris lumbricoides Tas virus
Distinguishing features
The integrated form of Ascaris lumbricoides Tas virus (AluTasV) is 7.6 kbp in length and consists of an internal domain flanked by two LTRs 256 bp in length. Insertions of AluTasV are flanked by 5 bp direct repeats derived from the insertion sites. There are approximately 50 copies of AluTasV distributed in the genome of A. lumbricoides. RNA transcripts and VLPs have not been observed but can be inferred based on the similarity in structure and coding capacity to that of other members of the family Metaviridae or the family Retroviridae. Reverse transcription is primed by tRNAarg which anneals to the pbs 6 bp downstream of the 5′-LTR. AluTasV encodes three overlapping ORFs: the first encodes the major structural protein and PR, the second ORF overlapping in the −1 frame encoding RT-RH and IN, and the third overlapping in the +1 frame encoding the env. The Env-like protein encoded by AluTasV contains a transmembrane domain but exhibits no sequence similarity with the env gene of Errantivirus or Retroviridae suggesting its independent acquisition. The likely origin of the AluTasV third ORF is the glycoprotein gB gene of herperviruses.
Species demarcation criteria in the genus
Members of this group have been identified in vertebrates, insects and nematodes. A reverse transcriptase phlylogenetic tree (Figure 2) indicates that all members of this group are well separated from members of the Metavirus and Errantivirus genera. Based on the sequence of the putative primer binding site, most viruses in this genus use either the acceptor stem of various tRNAArg or tRNAGly as the primer for minus-strand synthesis during reverse transcription. One continuous or two overlapping ORFs characterize the members of this group with the order of domains within the pol ORF (PR-RT-RH and IN) identical to that in the genera Metavirus and Errantivirus. Semotiviruses are particularly abundant in nematodes. The sequence of the Caenorhabditis elegans genome has revealed 13 families of viruses, but the majority of these are no longer active. An unusual feature of many of the C. elegans viruses is the presence of additional DNA between the 5′-LTR and the beginning of the first ORF. These additional sequences are variable within a family and completely different between families. One active group of sequences in the Caenorhabditis elegans Cer13 virus (CelCer13V) also contains a third Env-like domain between the pol encoded enzymatic domains and the 3′-LTR. This domain exhibits no sequence similarity with the domain in AluTasV, suggesting an independent acquisition. The likely origins of the CelCer13 Env-like domain is the G2 glycoprotein gene from phleboviruses. All individual species in the genus have less than 50% identity in their Gag protein sequences compared to all other species.
List of species in the genus Semotivirus
| Anopheles gambiae Moose virus |
|
|
| Anopheles gambiae Moose virus - Pink eye | [AF060859] | (AgaMooV-Pe) |
| Ascaris lumbricoides Tas virus |
|
|
| Ascaris lumbricoides Tas virus – al | [Z29712] | (AluTasV-al) |
| Bombyx mori Pao virus |
|
|
| Bombyx mori Pao virus – 703 | [L09635] | (BmoPaoV-703) |
| Caenorhabditis elegans Cer13 virus |
|
|
| Caenorhabditis elegans Cer13 virus - Bristol N2 | [Z81510] | (CelCer13V-BrN2) |
| Drosophila melanogaster Bel virus |
|
|
| Drosophila melanogaster Bel virus - white- zeste mottled | [U23420] | (DmeBelV-wzm) |
| Drosophila melanogaster Roo virus |
|
|
| Drosophila melanogaster Roo virus – roo | [AY180917] | (DmeRooV-roo) |
| Drosophila simulans Ninja virus |
|
|
| Drosophila simulans Ninja virus - white- chocolate | [D83207] | (DsiNinV-wc) |
| Fugu rubripes Suzu virus |
|
|
| Fugu rubripes Suzu virus – Fr | [AF537216] | (FruSuzV-Fr) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Semotivirus but have not been approved as species
None reported.
List of unassigned species in the family Metaviridae
None reported.
Phylogenetic relationships within the family
See Figure 2.
Similarity with other taxa
Like viruses of the family Pseudoviridae, the viruses of the family Metaviridae are clearly related to the viruses of the family Retroviridae. All of these families are related by reverse transcription and a viral core structure made up of Gag-like proteins. All members of the families Metaviridae, Pseudoviridae and Retroviridae also share the following: a proviral form characterized by LTRs, protease, RNase H and integrase activities essential for multiplication, read through-mediated (Gag-Pol) pol gene expression and tRNA primers (in some species). An important and somewhat controversial question is therefore the extent of the relationship of the members of the family Metaviridae to those of the family Retroviridae. Reverse transcriptase aa sequences are the most conserved sequences in retroelements and hence are the best character on which to base the phylogenetic relationship of these elements. Based on the phylogeny of their RT domains (Figure 2, see also Figure 4 in the chapter Pseudoviridae) members of the families Metaviridae and Pseudoviridae probably shared a common ancestor. The only major structural change between the genomes of members of these two groups was the movement of the IN domain from upstream to downstream of the RT-RH domain. The family Metaviridae is a numerous and diverse group of viruses distributed throughout eukaryotes. One lineage of the family Metaviridae in vertebrates appears to have given rise to the family Retroviridae.
The conversion of a lineage of the family Metaviridae into the family Retroviridae presumably occurred by the transduction of a gene encoding a ligand for cell-surface receptors or a cell fusion protein. The independent acquisition of a cell-surface receptor or fusion protein has occurred on at least five other occasions within the family Metaviridae. These include the Errantivirus genus in insects, Drosophila buzzatti Osvaldo virus (DbuOsvaV) and AthAthV within the genus Metavirus in insects and plants respectively, and AluTasV and CelCer13V within the genus Semotivirus in nematodes. The ease with which these viruses can gain, and presumably lose, an env-like gene means that this property is not always a reliable indicator of phylogenetic relationships. Finally, a second lineage in plants has become the family Caulimoviridae by the acquisition of a number of new genes that resulted in a number of changes to its life cycle (see description of Caulimoviridae).
Derivation of names
Erranti: from Latin errans, “to wander”.
Meta: from Greek metathesis for “transposition”. Also to connote some uncertainty as to whether these are true viruses or not.
Semoti: from Latin semotus, “distant, removed”. This prefix refers to the observation that based on the sequence of their RT domain, the viruses in this genus are distantly related to the other two genera of the family Metaviridae.
Further reading
Boeke, J.D. and Stoye, J.P. (1997). Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Varmus, H., Hughes, S. and Coffin, J., Eds), Retroviruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, pp 343-435.
Eickbush, T.H. and Malik, H.S. (2002). Evolution of retrotransposons. In: Craig, N., Craigie, R., Gellert, M. and Lambowitz, A., Eds), Mobile DNA II. American Society of Microbiology Press, Washington, DC, pp. 1111-1144.
Goodwin, T.J.D. and Poulter, R.T.M. (2001). The DIRS group of retrotransposons. Mol. Biol. Evol., 18, 2067-2082.
Malik, H.S., Henikoff, S. and Eickbush, T.H. (2000). Poised for contagion: evolutionary origins of the infectious abilities of insect errantiviruses and nematode retroviruses. Genome Res., 10, 1307-1318.
Marin, I. and Llorens, C. (2000). Ty3/Gypsy retrotransposons: description of new Arabidopsis thaliana elements and evolutionary perspectives derived from comparative genomic data. Mol. Biol. Evol., 17, 1040-1049.
Peterson-Burch, B.D., Wright, D.A., Laten, H.M. and Voytas, D.F. (2000). Retroviruses in plants? Trends Genet., 16, 151-152.
Contributed by
In reproducing this chapter in edited form from the Eighth ICTV Report, the editors gratefully acknowledge the contribution of the authors, Eickbush, T., Boeke, J.D., Sandmeyer, S.B. and Voytas, D.F.
Figures
Figure 1 Genome organization of representative members of the Metaviridae family. The integrated genome of each virus contains Long Terminal Repeats (LTRs) flanking a central sequence. Black boxes within the LTRs depict sequences repeated at the 5 and 3 ends of the virus transcripts (R regions). Open boxes below the viruses indicate gag, pol and env ORFs. Not all viruses within the genera Metavirus and Semotivirus encode an Env-like protein. Arrows indicate sites of ribosomal frameshifting. Conserved aa sequences in pol that identify protease (PR), integrase (IN) and reverse transcriptase/RNAse H (RT-RH) are labeled. Individual mRNAs are depicted below the ORF diagrams as arrows. Transcription of the DmeBelV and AluTasV has not been studied.
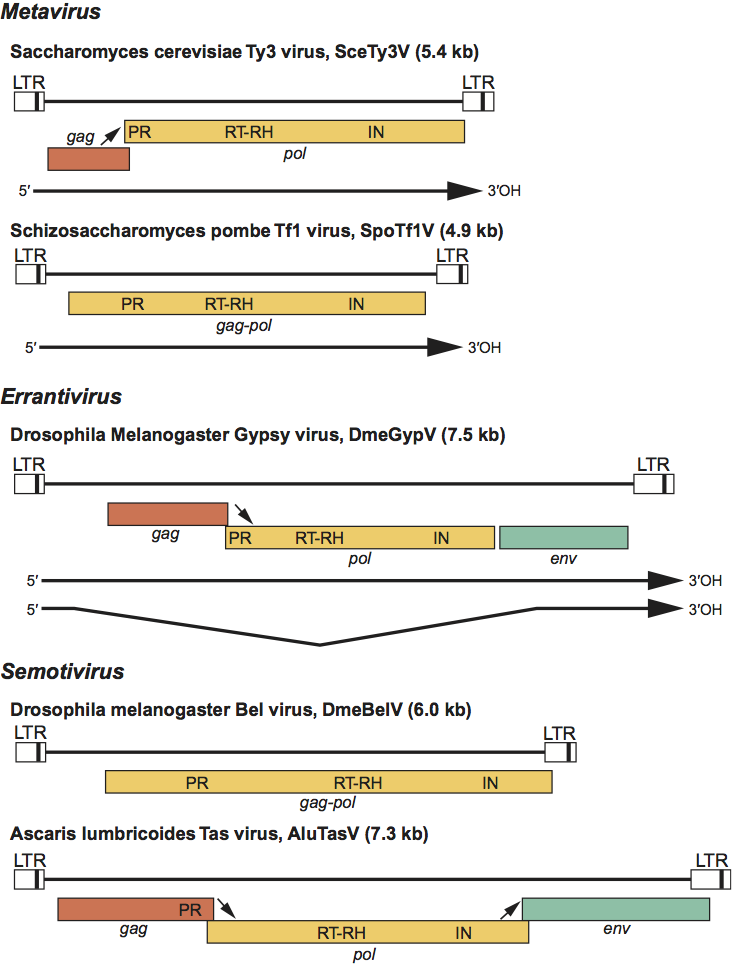
Figure 2 Phylogeny of the family Metaviridae and related groups based on their reverse transcriptase domain. The portion of the reverse transcriptase domain used in this analysis spans approximately 250 aa and includes the most conserved residues found in all retroelements. The phylogram is a 50% consensus tree of the viruses based on neighbor-joining distance algorithms and was rooted using sequences of members of the family Pseudoviridae. Bootstrap values (percentage of the time all elements are located on that branch) are shown for the major branches only. Viruses that are included in the various divisions of Metaviridae are indicated by the vertical lines to the right of each systematic name. Each group of viruses that are not part of the family Metaviridae is represented by a box with the length of the box related to the sequence diversity within that group. DIRS retrotransposons are mobile elements that utilize a reverse transcriptase closely related to that of the family Metaviridae but lack many structural features of this group and integrate by a different mechanism. The bar at the bottom represents divergence per site.