Family: Hepadnaviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Virion properties
Morphology
Hepadnaviruses are spherical, occasionally pleomorphic, 42–50 nm in diameter, with no evident surface projections after negative staining. Projections are visible in cryo-EM pictures (Figure 1C). The outer, detergent-sensitive, envelope contains the surface proteins and surrounds an icosahedral nucleocapsid core that is composed of one major protein species, the core protein. The nucleocapsid encloses the viral genome (DNA), the viral DNA polymerase, and associated cellular protein(s), including protein kinase and chaperones that appear to play a role in the initiation of viral DNA synthesis.
In the case of hepatitis B virus (HBV), the majority of nucleocapsid cores are about 36 nm in diameter and contain 240 core protein subunits (triangulation number T=4), while a minority are approximately 32 nm in diameter and consist of only 180 subunits (T=3). Hepadnavirus infection induces overproduction of surface proteins that are secreted into the blood as pleomorphic lipoprotein particles together with virus. In the case of HBV, these form 17–22 nm spherical particles and filaments (Figure 1).
Physicochemical and physical properties
The virion S20,w is approximately 280S. The buoyant density of virions in CsCl is approximately 1.25 g cm−3. Estimates of the buoyant density of particles lacking cores are 1.18–1.20 g cm−3. Virus-derived cores (lacking envelopes but containing nucleic acid) have densities of approximately 1.36 g cm-3.
Nucleic acid
The genome consists of a partially dsDNA that is held in a circular conformation by base pairing in a cohesive overlap between the 5’ ends of the two DNA strands. The length of the cohesive overlap is about 240 bp for the orthohepadnaviruses and 50 bp for the avihepadnaviruses. The size of the genome ranges from 3.0 to 3.3 kb in different family members; the viral DNA has an S20,w of about 14 and a G+C content of about 48%. One strand (negative sense, i.e. complementary to the viral mRNAs) is full-length, whereas the other varies in length. For both the orthohepadnaviruses and the avihepadnaviruses, the negative strand DNA has an 8–9 nt terminal redundancy. The 5’ end of the negative strand DNA is covalently attached to the terminal protein (TP) domain of the viral DNA polymerase, and the 5’ end of the positive sense DNA has a covalently attached 19 nt, 5’ capped oligoribonucleotide primer. The 3’ end of the positive strand terminates at a variable position in different molecules, creating a single stranded gap that may account for 60% of the HBV genome but is usually very short in avihepadnaviruses.
Proteins
Virions and empty subviral particles may contain two or three surface proteins, with a common C-terminus but distinct N-termini due to different sites of translation initiation. Typically, virions contain a small transmembrane surface protein (SHBs), in the orthohepadnaviruses an intermediate size (MHBs) and a large (LHBs) protein that is myristylated at the N-terminus. In many cases, more than one form of each of the above proteins occurs due to alternative patterns of glycosylation. For HBV and WHV, virions and filaments are enriched in L proteins and empty spheres consist predominantly of S proteins, while for duck hepatitis B virus (DHBV), L and S proteins are distributed evenly between particle types.
The core protein has a large N-terminal domain and a small RNA-binding domain at the C-terminus. Core protein above a threshold concentration can self-assemble via dimers to complete nucleocapsids in the absence of other viral components.
The polymerase protein consists of an N-terminal domain (TP) with a DNA primer function, a spacer region of variable size, a reverse transcriptase and an RNase H domain. The TP domain is covalently attached to the 5’-end of the minus strand of viral DNA via a tyrosine residue, which serves as the primer for initiation of reverse transcription.
Orthohepadnaviruses contain a fourth ORF (“X” gene) situated downstream from the S gene and partly overlapping the cohesive 5’-terminal region. This codes for a non-structural protein that can function as a promiscuous transcriptional activator and, for the woodchuck hepatitis virus (whv), it has been shown to be required for efficient in vivo replication. Avihepadnaviruses have an ORF in a similar location, but it remains unclear whether it has a role in infection. At high expression levels in cell culture systems, the X proteins induce apoptosis.
Host proteins contained within nucleocapsids include heat shock protein Hsp 70 and heat shock protein Hsp 90, which, at least in the case of duck hepatitis B virus (DHBV), appear to be part of a multicomponent chaperone complex involved in replication and nucleocapsid assembly. A protein kinase has been detected in HBV nucleocapsids.
The core protein also exists in a secreted soluble form (“e” antigen) that is translated from an additional start codon 29 codons upstream of the core start codon. This additional precore sequence functions as a signal peptide. The “e” antigen is not essential but is conserved in all hepadnaviruses and seems to modulate the immune response.
Lipids
Lipid constitutes 30–40% of the viral envelope or of the empty particles. It is derived from a host membrane compartment intermediate between the ER and Golgi, and includes phospholipids, cholesterol, cholesterol esters and triglycerides.
Carbohydrates
Demonstrated in particles and virions of orthohepadnaviruses as N-linked glycans of the complex types. Many virus isolates also contain O-linked glycans in the M surface protein.
Genome organization and replication
The hepadnavirus genome contains the following major ORFs; precore/core (preC/C), polymerase (P), env or surface (preS/S), and in the case of orthohepadnaviruses, a fourth ORF, the X gene (Figure 2).
The preC/C ORF codes for two distinct products: one is the core protein forming the protein shell of the nucleocapsid, the other, made by translation of the joint preC/C ORF, is the precore protein which is targeted into the cell’s secretory pathway, processed at both ends and eventually found in the serum of infected individuals as e antigen. Both products are translated from genomic, terminally redundant, polyadenylated 3.5 kb transcripts with slightly different 5’-ends. The longer precore mRNAs contain the preC initiation codon, whereas the shorter core mRNA lacks it. The P ORF covers some 80% of the genome and encodes the viral replication enzyme P, which is also an indispensable component in the assembly process (see below). P protein is translated from the same genomic RNA that directs synthesis of core protein by internal initiation. The env or surface gene consists of three in-phase ORFs, termed in 5’- to 3’-direction, preS1, preS2 and S. S can be expressed separately to give the small or S protein; cotranslation of preS2/S yields the middle or M protein (orthohepadnaviruses), and that of the entire preS1/preS2/S gene the large or L protein. Thus, the S domain is common to all three forms of surface proteins. As for the preC/C ORF, this is achieved by the generation of mRNAs with staggered 5’ ends in which the initiator codons of the preS1, the preS2 or the S region are the first to be encountered by translating ribosomes. L protein is translated from a 2.4 kb mRNA, and M and S from a set of 2.1 kb transcripts. All viral transcripts are 3’-terminally colinear, ending after a unique polyadenylation signal located in the C gene. The X gene encodes a pleiotropic transcriptional activator that appears to be required for establishment of a normal infection with WHV, and has been implicated in one proposed mechanism for hepadnavirus carcinogenesis. The DNA sequence of HBV has two enhancer regions (ENHI and ENHII), a negative regulatory element NRE, four promoters (preC/C, preS1, preS2/S and X), two 11-base direct repeat sequences (DR1 and DR2), a polyadenylation signal (TATAAA), and putative glucocorticoid-responsive elements (GRE). The 5’ end of the negative strand is located within DR1, and the 5’-end of the positive strand is at the 3’ boundary of DR2.
Replication can be considered in two stages: an incoming or afferent arm in which the input viral genome enters the nucleus and is converted to covalently closed circular (ccc) DNA (cccDNA), and an outgoing or efferent arm in which RNA transcripts from the cccDNA are encapsidated and reverse transcribed within core particles in the cytoplasm, and the resulting genomic DNA is either transported to the nucleus or enveloped and secreted (Figure 3).
There is evidence that the infectious DNA-containing virion binds to its target cell via interaction of the L protein with cellular receptor(s) that are not yet fully characterized. The nucleocapsid is presumably delivered to the cytoplasm and transported to the nuclear pore where the genome is released to the nucleoplasm. Repair of the single stranded gap is carried out, though it remains uncertain whether this is achieved by the viral DNA polymerase or a host polymerase. Removal of the TP and oligoribonucleotide from the negative and positive strands, respectively, takes place, followed by DNA ligation, converting the viral genome to cccDNA. These steps are believed to involve the action of cellular enzymes. cccDNA, in the form of a histone-associated minichromosome, provides a stable template for transcription.
Genomic and sub-genomic RNAs (sgRNAs) are transcribed by host RNA polymerase II into a number of RNA size classes, some of which also show microheterogeneity at the 5’ end but all terminate at a common 3’-polyadenylation site. The RNAs of HBV and WHV contain a post-transcriptional regulatory element PRE, which allows for cytoplasmic transport without splicing. The largest of these RNAs is translated to form precore protein. A slightly shorter RNA encodes the core protein and (by internal initiation) the polymerase protein, and also serves as the template for reverse transcription. The polymerase protein, along with host chaperone proteins, associates with a specific encapsidation signal (ε), which is closed to the 5’ end of the pregenomic RNA. For efficient minus strand DNA synthesis epsilon has to interact with the 19 nt long phi signal, which locates 32 nt upstream of DR1 at the 3’ end of the RNA and this preassembly complex apparently triggers assembly of core protein dimers into complete nucleocapsids.
Concurrently, the different classes of sgRNAs are translated to produce the various surface gene products (L, M and S), which oligomerize and bud into the lumen of a post ER/pre-Golgi compartment to give rise to both empty subviral particles and virions.
Reverse transcription of pregenomic RNA takes place within cytoplasmic immature cores. This process uses the TP domain of polymerase as primer for first strand synthesis, and a short, undigested, capped oligoribonucleotide derived from the 5’ end of the template RNA and extending through the proximal copy of DR1 as the second strand primer. Synthesis of both strands requires transfer reactions. Reverse transcription initiates with the copying of 4 nt from a bulge in . This product is then annealed to a complementary sequence at the 3’ copy of DR1, and it is from this site that synthesis of the full-length minus strand progresses. Plus strand synthesis involves a transfer of the RNA primer from the 3’ end of the minus strand to a remote site, DR2, which is near the 5’ end of the minus strand and identical in sequence to DR1. It is here that plus strand synthesis normally begins. Plus strand elongation requires a second translocation, from the 5’ to the 3’ end of the minus strand template, to form an open circular genome with a less than full-length (+) strand, maintained by overlapping cohesive ends. Nucleocapsids containing partly reverse transcribed DNA that have associated with cytoplasmically located preS domains of the L envelope protein may then bud into the lumen of multivesicular bodies as maturing virions, or alternatively may be transported to the nucleus, thereby increasing the pool of cccDNA. Although integration of viral DNA into the host genome is not required for replication and appears to be an infrequent event, integrated viral DNA, often containing deletions, inversions and duplications, is found in hepatocellular carcinoma (HCC) cells in culture and in patients, as well as in apparently normal livers from chronic carriers. An aberrant linear viral DNA, formed when plus strand synthesis initiates from an untranslocated primer, appears to be the precursor to the majority of integration events.
Antigenic properties
Three principal antigens have been identified for hepadnaviruses, designated surface, core and e antigen. These are abbreviated HBsAg, HBcAg, HBeAg for the HBV-related antigens, DHBsAg, DHBcAg and DHBeAg for DHBV-related antigens, etc., while the corresponding antibodies are designated anti-HBs, anti-HBc, anti-DHBs, anti-DHBc, etc. HBsAg is involved in neutralization. It cross-reacts to a limited extent with the analogous antigens of WHV and ground squirrel hepatitis virus (GSHV), but not with DHBsAg. PreS antigens and the HBsAg loop in the S protein bear specific neutralization determinants. S proteins are sufficient to stimulate protective immunity.
HBeAg and HBcAg proteins share common sequences and epitopes, but also contain epitopes that distinguish these two proteins from each other. HBeAg is a 16 kDa truncated derivative of HBcAg. It is found as a soluble antigen in the serum of patients. HBcAg has been found to cross-react more strongly with the WHV core antigen than is seen between the corresponding surface antigens. In much of the earlier literature, the term surface antigen or HBsAg is used arbitrarily to refer to either the antigenic specificity, various protein products of the preS1/preS2/S gene, or the empty 17–22 nm HBsAg-bearing particles. The term “antigen” should not be used if “protein” or “particle” is intended. Similar considerations apply to the use of “core antigen”.
Biological properties
All hepadnaviruses show narrow host specificity. In vitro, replication of many hepadnaviruses has only been demonstrated following transfection of tissue culture cells by cloned viral DNA, resulting in the production of infectious virus. Replication of several hepadnaviruses has been achieved following inoculation of primary hepatocytes with serum that contains virus.
Hepadnavirus infections in vivo possess characteristic features:
- They are markedly hepatotropic, although viral antigens and nucleic acids can also be detected in white blood cells (and in some extra-hepatic sites, e.g. pancreas, spleen and kidney with avihepadnaviruses).
- Infection may be transient or persistent, the outcome depending on factors such as host age and dose of inoculum. Persistent infection is more common in neonates and in immuno-compromised hosts. Persistent infections are typically life-long and can be accompanied by high levels of virions and subviral particles in the circulating blood.
- Empty subviral particles, composed of excess virus envelope material, are present in much greater numbers than complete virions in most individuals and at most stages of infection.
- Virus replication is generally thought to be non-cytopathic, and different degrees of ongoing liver damage in different individuals are thought to be governed by different degrees of immune-mediated damage to infected hepatocytes.
- In ortho-, but not avihepadnavirus infections, persistent virus infection confers a significantly increased rate of development of primary hepatocellular carcinoma, and a number of direct and indirect mechanisms have been described.
Genus Orthohepadnavirus
Type species Hepatitis B virus
Distinguishing features
Viruses of this genus infect mammals, with a narrow host range for each virus species. The only known natural host of HBV are humans and greater apes (chimpanzees, gorillas, orangutans and gibbons). Virions of HBV are 40–45 nm in diameter with a 32–36 nm internal nucleocapsid, and subviral HBV particles are typically spherical (16–25 nm diameter) and filamentous (20 nm diameter and variable in length). The genome of HBV is 3.2 kb with a cohesive overlap of 240 bp. The viruses have an S protein of approximately 226 amino acid residues (aa) as a major envelope protein, an M protein of about 271 aa (which appears unnecessary for infection in experimental situations) and an L protein of about 400 aa.
The envelope, or surface, proteins are partially glycosylated, thus generating doublets in gel electrophoresis, e.g. for HBV, P24/GP27 for S, P39/GP42 for L, and, in the case of M, GP33/GP36 due to an additional glycosylation in the preS2 sequence. The HBV core protein is approximately 180 aa, and the virus encodes an HBx protein of 154 aa whose natural function in the virus life cycle is uncertain.
At least five antigenic specificities have been identified for HBsAg. A group determinant (a) is shared by virtually all HBsAg preparations. Mutations in this region have been found in immunized individuals who subsequently became infected, in HBV carriers, particularly in occult HBV infection without detectable HBsAg, and in infected individuals given immunotherapy. Two pairs of subtype determinants (d,y and w,r) have been demonstrated that are generally mutually exclusive (and thus usually behave as alleles). Antigenic heterogeneity of the w determinant, and additional determinants such as q, x or g have also been described. Thus, eight major serological subtypes are found (ayw, ayw2, ayw3, ayw4, ayr, adw2, adw4 and adr); they have distinct geographical distributions with some overlap. DNA sequence analysis has now replaced antigenic typing in defining viral genotypes and has distinguished genotypes, or clades, that differ between each other by 8–14% at the nucleotide level. Different genotypes also have different geographical distributions, and there is some, but not complete, correspondence between genotype and serological subtype. HBV strains infecting apes fall into species-specific genotypes, which have been suggested to reflect co-evolution of virus and host and not horizontal transmission between primate species.
Woolly monkey hepatitis B virus (WMHBV) is closely related in DNA sequence to HBV, with 20% sequence variation, but, unlike HBV, preferentially infects the woolly monkey and can be transmitted to the spider monkey; it transmits only poorly to the chimpanzee, which is highly susceptible to human HBV genotypes.
Different isolates of WHV show <3.5% nucleotide sequence variation. A virus of arctic ground squirrels (Arctic squirrel hepatitis virus, ASHV) differs from WHV and GSHV to about the same extent (about 15% nt changes) as these two latter viruses differ from each other.
Biological properties
HBV may cause as a consequence of the host immune response to infection, acute and chronic hepatitis, and immune complex diseases like periarteritits nodosa, glomerulonephritis, and infantile papular acrodermatitis. Late sequelae are liver cirrhosis and hepatocellular carcinoma. An asymptomatic carrier state with high viremia may develop, particularly after perinatal infection or under immune suppression.
Horizontal transmission of HBV usually occurs by: (i) percutaneous contact with infected blood or body fluids, e.g. intravenous drug abuse or use of infected blood or blood products; (ii) sexual contact; (iii) perinatal transmission from an infected mother; and (iv) “inapparent horizontal” transmission, particularly between children in low socio-economic communities, thought to be due, at least in part, to unrecognized exposure to open skin breaks or mucous membranes. In communities with a high prevalence of infection, routes (iii) and (iv) predominate, while in low prevalence communities, infections are acquired later in life and involve particularly routes (i) and (ii).
Hepatitis occurs in woodchucks and squirrels infected with their respective viruses, and chronic infection leads to a risk of hepatocellular carcinoma even greater than that in chronic carriers of HBV. In the case of WHV, hepatocellular carcinoma frequently occurs within 2 years of infection.
Woolly monkey hepatitis B virus causes hepatitis in its host, but is not yet known to have a role in liver cancer.
Species demarcation criteria in the genus
The species demarcation criteria in the genus are:
- Nucleotide sequence diversity; WHV/HBV 40%; GHSV/WHV 15%; WMHBV/HBV 20%; WMHBV/WHV 30%.
- Differences in host range: HBV infection is limited to primates, but HBV may infect primary hepatocyte cultures from Tupaia belangueri which is not a primate. GSHV infection has been experimentally transferred to chipmunks and woodchucks but not to several related ground squirrel species; WHV also has a narrow host range, being reported not to infect ground squirrels or other rodent species. WMHBV is transmitted to the spider monkey.
- Oncogenicity: HBV, WHV and GSHV have been associated with primary liver cancer in infected hosts. However, the proposed mechanisms are different in each case, and incidence and typical time scales differ, being highest with WHV and lowest with HBV.
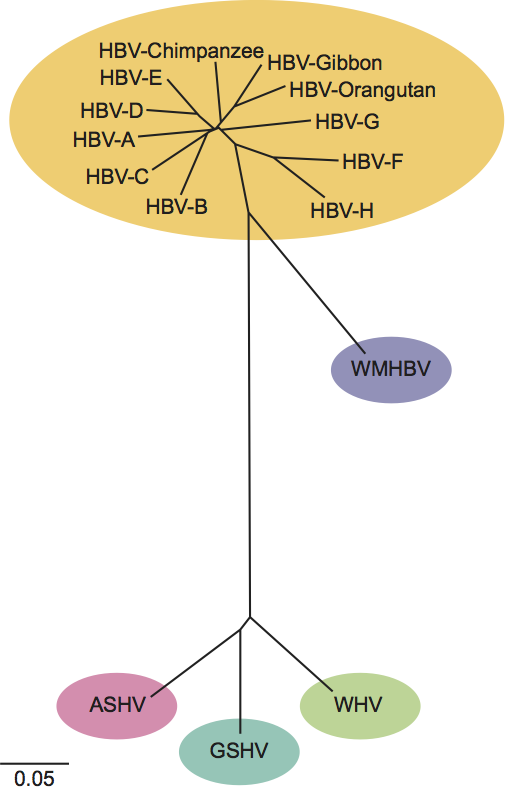
A number of related viruses that are strong candidates for inclusion in this genus have been isolated from non-human primates (chimpanzees, gibbons, orangutan and gorilla) and from various rodent species (Arctic ground squirrel and Richardson’s ground squirrel). As illustrated in Figure 4, these isolates are quite similar to assigned isolates of HBV, WHV or GSHV.
List of species in the genus Orthohepadnavirus
| Ground squirrel hepatitis virus |
|
|
| Ground squirrel hepatitis virus | [K02715] | (GSHV) |
| Hepatitis B virus |
|
|
| Hepatitis B virus - A | [X02763] | (HBV-A) |
| Hepatitis B virus - B | [D00330] | (HBV-B) |
| Hepatitis B virus - C | [AY123041] | (HBV-C) |
| Hepatitis B virus - D | [V01460] | (HBV-D) |
| Hepatitis B virus - E | [X75657] | (HBV-E) |
| Hepatitis B virus - F | [X69798] | (HBV-F) |
| Hepatitis B virus - G | [AF160501] | (HBV-G) |
| Hepatitis B virus - H | [AY090454] | (HBV-H) |
| Hepatitis B virus - gib I | [AJ131569] | (HBV-gib I) |
| Hepatitis B virus - gib II | [AJ131571] | (HBV-gib II) |
| Hepatitis B virus - gib III | [AJ131572] | (HBV-gib III) |
| Hepatitis B virus - gib IV | [AJ131573] | (HBV-gib IV) |
| Hepatitis B virus - gib V | [AJ131574] | (HBV-gib V) |
| Hepatitis B virus - orangutan | [AF193864] | (HBV-orangutan) |
| Hepatitis B virus - chHBV | [D00220] | (chHBV) |
| Woodchuck hepatitis virus |
|
|
| Woodchuck hepatitis virus | [J02442] | (WHV) |
| Woolly monkey hepatitis B virus |
|
|
| Woolly monkey hepatitis B virus | [AF046996] | (WMHBV) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Orthohepadnavirus but have not been approved as species
| Arctic squirrel hepatitis virus | [U29144] | (ASHV) |
Phylogenetic relationships within the genus
See figure 4.
Genus Avihepadnavirus
Type species Duck hepatitis B virus
Distinguishing features
DHBV virions are spherical, 46–48 nm in diameter, with a nucleocapsid that is 35 nm in diameter and exhibits projections. Empty particles composed of excess envelope material are pleomorphic and up to 60 nm diameter. The single stranded gap in the virion DNA is usually very short, at about 12 nt. DHBV lacks an X gene containing a conventional initiation codon, but some other avihepadnaviruses may have an X gene with a regular start codon. Virus particles have only the largest (36 kDa) and smallest (17 kDa) S proteins. Transmission is predominantly vertical. Heron hepatitis B virus (HHBV) differs from DHBV in that a highly conserved ORF is present upstream of C in a position analogous to the X gene of orthohepadnaviruses.
Biological properties
DHBV is maintained in domestic duck flocks through vertical transmission from viremic ducks. The virus infects the developing liver in ovo and is not recognized sufficiently by the host immune response to produce hepatitis and liver disease, or to eliminate the virus. Liver cancer has not been associated with chronic infection. Transmission to neonates may also occur, leading to chronic infection. Transmission to adults generally leads to transient infection. The biology of HHBV infections in its natural host has not been studied.
Species demarcation criteria in the genus
The species demarcation criteria in the genus are:
- Nucleotide sequence diversity: HHBV/DHBV 21.6%
- HHBV can be transmitted to herons but not to ducks.
A number of other characterized viruses have been isolated from geese and ducks, with sequences reportedly more closely related to that of DHBV than HHBV. A virus isolated from grey crowned cranes (Balearica regulorum), designated crane hepatitis B virus (CHBV), has sequences more closely related to DHBV than HHBV, and has been shown to infect primary duck hepatocytes. A virus closely related to HHBV has been isolated from white storks and designated stork hepatitis B virus (STHBV). Like HHBV, STHBV has a low infectivity for duck hepatocytes. Tentatively, STHBV may be assigned a variant of HHBV.
List of species in the genus Avihepadnavirus
| Duck hepatitis B virus |
|
|
| Duck hepatitis B virus | [K01834] | (DHBV) |
| Ross’ goose hepatitis B virus | [M95589] | (RGHBV) |
| Crane hepatitis virus | [AJ44111] | (CHBV) |
| Heron hepatitis B virus |
|
|
| Heron hepatitis B virus | [M22056] | (HHBV) |
| Stork hepatitis B virus | [AJ251937] | (STHBV) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Avihepadnavirus but have not been approved as species
None reported.
Phylogenetic relationships within the family Hepadnaviridae
Orthohepadnaviruses and avihepadnaviruses are distinguished by the following criteria:
- Low nucleotide sequence identity
- Differences in genome size (about 3.2 kb for orthohepadnaviruses and 3.0 kb for avihepadnaviruses)
- Larger core proteins and no M surface protein for the avihepadnaviruses
- Host range restricted to either mammals or birds, respectively.
Similarity with other taxa
Reverse transcription, as an essential step in replication, is a common feature of hepadnaviruses, retroviruses and caulimoviruses. Hepadnaviruses and retroviruses also contain three major genes, each with the same function and in the same order (i.e. core-polymerase-pre S/S and gag-pol-env respectively). A fundamental distinction is that, with hepadnaviruses, the form of the genome in extracellular virions is DNA and reverse transcription takes place during the efferent or outgoing arm of the replication cycle, whereas the reverse holds true for retroviruses (with the exception of the spumaviruses, in which some infectious particles appear to contain a DNA genome). Retroviruses use tRNAs as primers for the DNA minus strand, whereas hepadnaviruses utilize a tyrosine in the polymerase itself. The polymerase protein of hepadnaviruses does not contain a protease or integrase function. Many other aspects are distinctly different in both virus families, partly due to the extremely small size of the hepadnaviral genome and the need to exploit this restricted genetic space efficiently by using the considerable overlap of both coding regions and regulatory elements.
Derivation of names
Avi: from Latin avis, “bird”.
DNA: deoxyribonucleic acid.
Hepa: from Greek hepar, “liver”.
Ortho: from Greek orthos, “straight”.
Further reading
Ganem, D. and Schneider, R.J. (2001). Hepadnaviridae: The viruses and their replication. In: Knipe, D.M. and Howley, P.M. (Eds), Fields Virology, 4th edn. Lippincott, Williams & Wilkins, Philadelphia, pp. 2923-2969.
Grethe, S., Heckel, J.-O., Rietschel, W. and Hufert, F.T. (2000). Molecular epidemiology of hepatitis B virus variants in nonhuman primates. J. Virol., 74, 5377-5381.
Hollinger, F.B. and Liang T.J. (2001). Hepatitis B virus. In: Knipe, D.M. and Howley, P.M. (Eds), Fields Virology, 4th edn. Lippincott, Williams & Wilkins, Philadelphia,pp. 2971-3036.
Kann, M. and Gerlich, W.H. (1998). Hepatitis B. In: Collier, L., Balows, A. and Sussman, M. (Eds), Topley and Wilson's Microbiology and Microbial Infections, 9th edn. Arnold, London, pp. 745-774.
Liang, T.J. (2009). Hepatitis B: the virus and disease. Hepatology, 49, S13-21.
Nassal, M. (2008). Hepatitis B viruses: reverse transcription a different way. Virus Res., 134, 235-249.
Newbold, J.E., Xin, H., Tencza, M., Sherman, G., Dean, J., Bowden, S. and Locarnini, S. (1995). The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J. Virol., 69, 3350-3357.
Prassolov, A., Hohenberg, H., Kalinina, T., Schneider, C., Cova, L., Krone, O., Frölich, F., Will, H. and Sirma, H. (2003). New hepatitis B virus of cranes that has an unexpected broad host range. J. Virol., 77, 1964-1976.
Schaefer, S. (2007). Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J. Gastroenterol., 13, 14-21.
Simmonds, P. (2001) The origin and evolution of hepatitis viruses in humans. J. Gen. Virol., 82, 693-712.
Contributed by
Mason, W.S., Gerlich, W.H., Taylor, J.M., Kann, M., Mizokami, T., Loeb, D., Sureau, C., Magnius, L. and Norder, H.
Figures
Figure 1 (A) Atomic resolution rendering of a particle of hepatitis B virus capsid (HBV) (Wynne, S.A., Crowther, R.A. and Leslie, A.G. (1999). Mol. Cell., 3, 771780). (B) Diagram representing the T=4 structure of an HBV core particle. (C) High resolution cryo-electron micrograph of normal (4252 nm) isometric virus with icosahedral capsid or core surrounded by a coat of HBsAg, and of smaller (ca. 22 nm) spheres and rods composed of viral envelope proteins. The bar represents 65 nm. (Courtesy of B. Boettcher, J. Monjardino and R.A. Crowther.) (Bottom). Negative contrast electron micrographs of HBV virions (left) and virus-associated particles (centre and right), together with an SDS-PAGE protein profile of each particle form to the left of the relevant micrograph. LHBs, MHBs and SHBs refer to large, middle and small HB surface proteins, respectively. HBc, hepatitis B core proteins. GP, glycoprotein; P, protein. The identities of the slower migrating bands are unknown.
(Courtesy of W. Gerlich.)
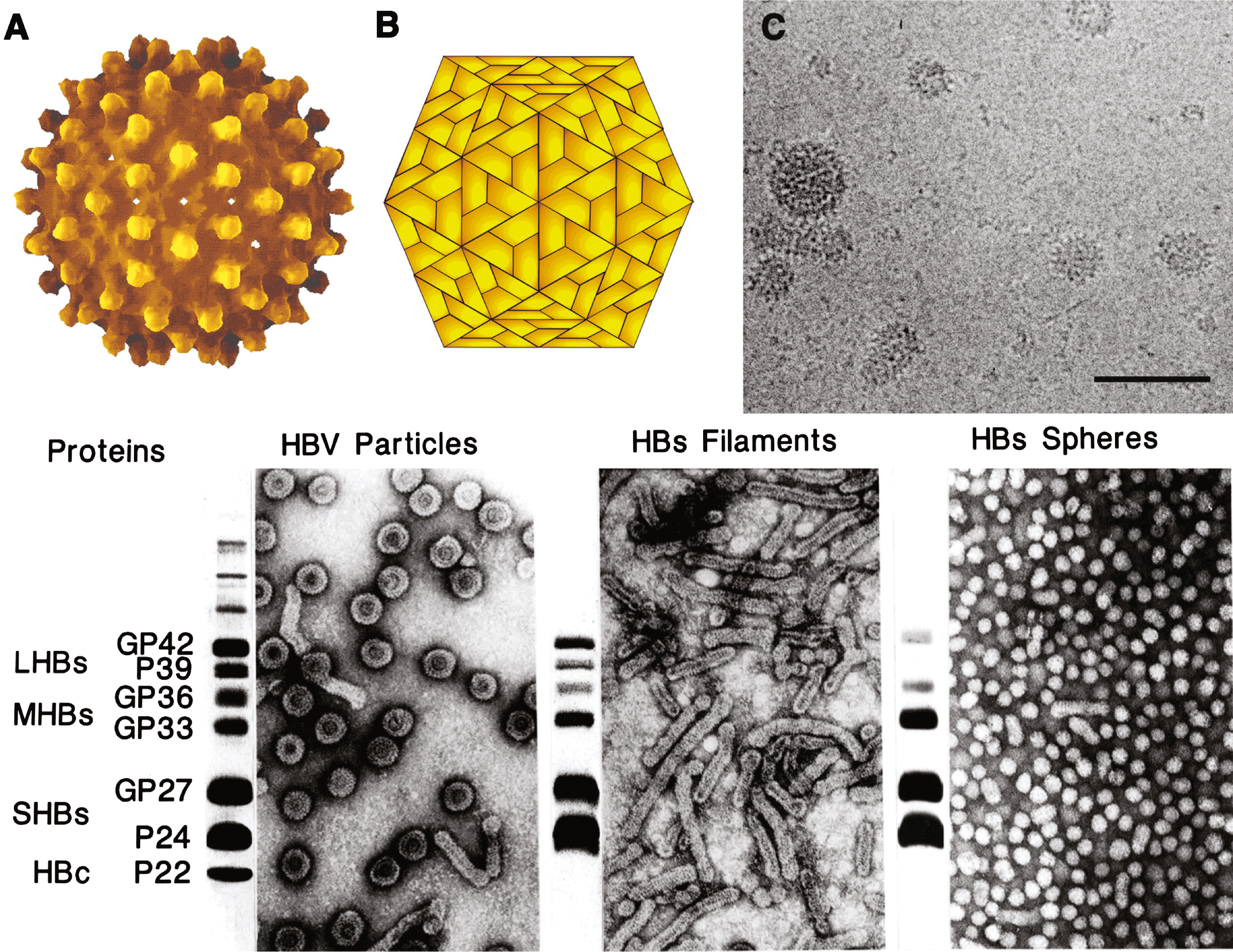
Figure 2 Genome organization and regulatory elements of orthohepadnaviruses are shown for a typical HBV isolate of genotype A. The outer circle represents the structure of relaxed circular, viral DNA found within virions, while the inner circle illustrates the structure and regulatory elements on cccDNA, the covalently closed circular DNA from which viral mRNAs are transcribed in the nucleus of the infected cell (red = positive strand; blue = negative strand). Numbering starts at the unique EcoRI restriction site located approximately at the junction of the preS1 and preS2 domains in the ORF for the viral envelope proteins. The regulatory elements on the DNA are depicted at their approximate positions. The promoters (P) are shown as grey boxes, and the enhancers (Enh), a glucocorticoid responsive element (GRE), and a CCAAT element (CCAAT) are depicted as black boxes. The basal core promoter is regulated by the negative regulatory element (NRE, not shown), which overlaps with Enh II. Liver-specific promoters are drawn in light grey; non-tissue-specific promoters are depicted as medium grey boxes. The ORFs are drawn as arrows with their corresponding start and termination sites. The viral mRNAs are depicted as black circles in the middle region. The black triangles represent their 5 ends; the 3 end is common and linked to an approximately 300 nt polyA tract. The regulatory elements on the RNAs are depicted as a red box (encapsidation signal ), a black box (polyadenylation signal), in pink (DR1), in blue (phi) and in light blue (posttranscriptional regulatory element [PRE]). The genomic DNA is depicted as it is found in the virion. The minus DNA strand is drawn as a blue line with its terminal redundancy (r). The polymerase (green oval) is linked to the 5 end of the minus strand. The plus-strand DNA is shown as a red line. The dotted red line represents the variation of the 3 end of the plus strand DNA. The 5 end of the plus strand is bound to its capped RNA primer, depicted as a black, wavy line. The dotted grey line between the polymerase and the 3 end of the plus strand DNA reflects the fact that the polymerase is bound to the 5 end of the minus strand DNA, but interacts with the variable 3 end of the plus strand DNA for its elongation. The regulatory elements on the minus strand DNA are the DR2 (red box) and the M, 5E and 3E elements, which are required for circularization of the genome. Note that their position and size are approximate, since these elements are not yet completely characterized.
(From Kann, M. (2002). Structure and molecular virology. In: Hepatitis B virus Human Virus Guide. (S. Locarnini and C.L. Lai, Eds.), ch 2. International Medical Press, London; with permission.)
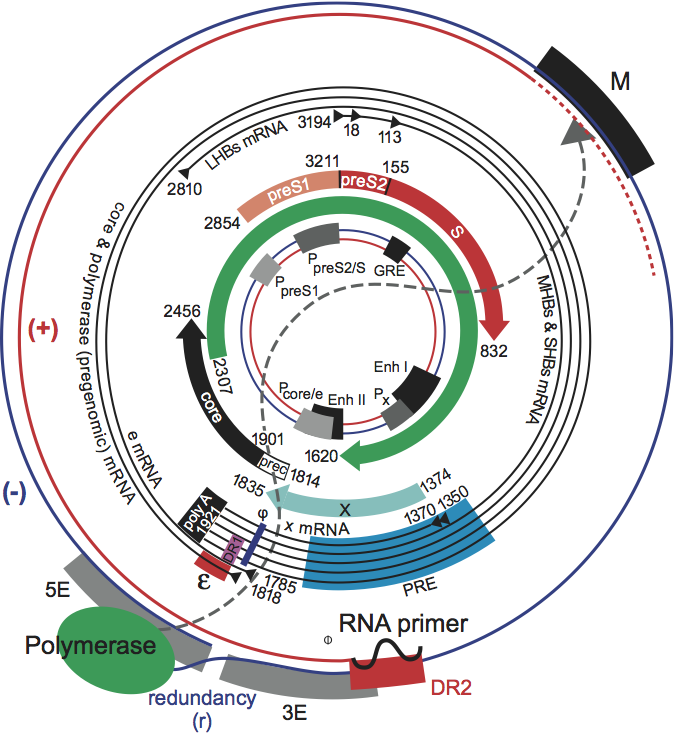
Figure 3 Hepadnavirus replication strategy. For details see text.
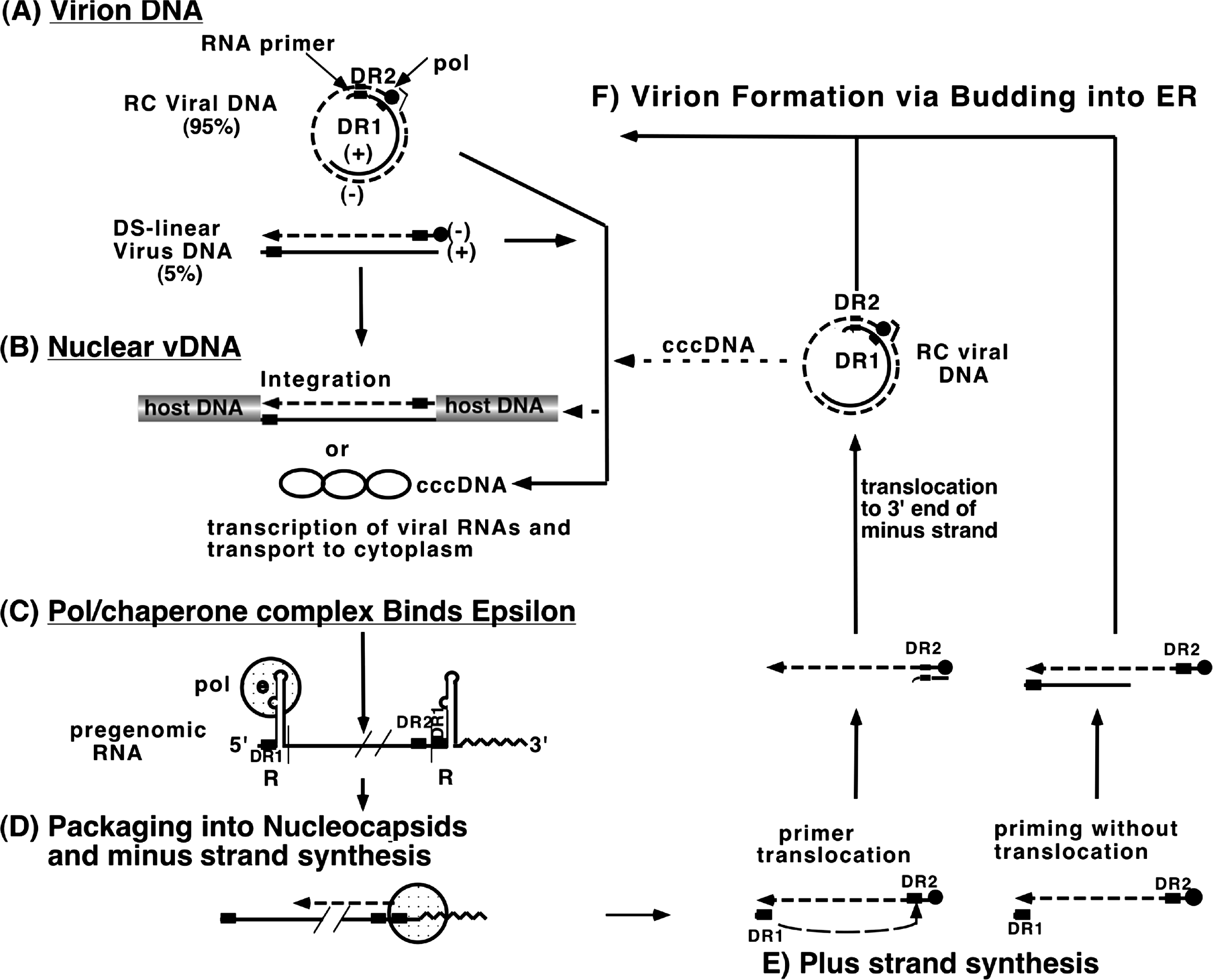
Figure 4 Phylogenetic tree of the genus Orthohepadnavirus. Complete genomes of hepatitis B virus (HBV) genotypes A (X02763), B (D00330), C (M12906), D (J02203), E (X75657), F (X69798), G (AF160501) and H (AY090454), and isolates found in chimpanzee (D00220), orangutan (AF193864) and gibbon (U46935), were aligned using Clustal W with orthohepadnavirus genomes from woolly monkey hepatitis B virus (WMHBV) (AF046996), woodchuck hepatitis virus (WHV) (J02442), ground squirrel hepatitis virus (GSHV) (K02715) and Arctic squirrel hepatitis virus (ASHV) (U29144). The alignment was tested with the neighbor-joining method. Calibration bar: substitutions per site.
(Courtesy of Schaefer.)